All published articles of this journal are available on ScienceDirect.
A Comparative Analysis of the Spanish Flu 1918 and COVID-19 Pandemics
Abstract
Two devastating pandemics, the Spanish Flu and COVID-19, emerged globally in 1918 from America and 2019 from China, respectively. Influenza virus A H1N1, which caused Spanish Flu and SARS-CoV2, which caused COVID-19, belong to different virus family and bear different structure, genomic organization and pathogenicity. However, the trajectory of the current outbreak of COVID-19 depicts a similar picture of the Spanish Flu outbreak. Estimates suggest that ~500 million infected cases and ~50 million deaths occurred globally from 1918-1919 due to the H1N1 virus. While SARS-CoV2 accounted for ~2 million cases and 130,885 deaths just within three and a half months, and the number is still increasing. To contain the spread of COVID-19 and to prevent the situation which happened a century back, it becomes essential to examine and correlate these pandemics in terms of their origin, epidemiology and clinical scenario. The strategies tailored to control the Spanish Flu pandemic may help to contain the current pandemic within time.
1. INTRODUCTION
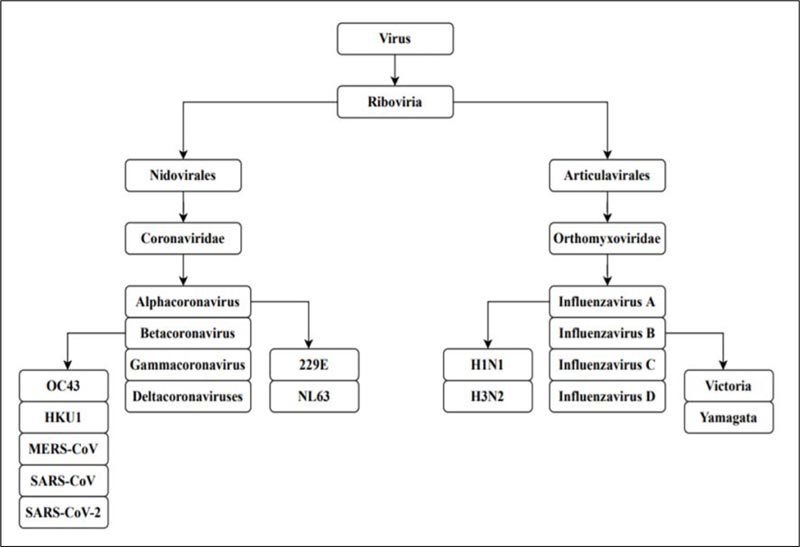
A pandemic is an extensive outbreak of a contagious disease, significantly increasing the probability of morbidity and mortality over a wide range of geographical area [1]. The severity of the pandemic is generally high as it affects a substantial volume of society due to the nonexistence of immunity against the causative agent [2]. The world witnesses pandemics mostly caused by viruses having a high impact on civilization. The current outbreak of the Severe Acute Respiratory Syndrome Coronavirus 2 [SARS-CoV-2] has caused substantial crisis across the globe following a more severe route compared to the other viruses of its family [3]. The Spanish flu pandemic of 1918 is regarded as the most deadly pandemic the world has faced since 1900 [4]. Such disease outbreaks globally present a great menace to global health. The Spanish Flu pandemic was caused by Influenza virus A H1N1, which belongs to the family Orthomyxoviridae, while SARS-CoV-2 of family Coronaviridae is responsible for the COVID-19 pandemic [5,6] (Fig. 1).
The origin, spread and the reason for occurrences of novel viruses responsible for a major crisis like pandemics are still unknown. Though the H1N1 and SARS-CoV-2 belong to a different family and possess various characteristics, a comprehensive investigation, comparing their trajectories, may aid us in understanding the virus pandemics in a better way. We also may use the incidences from 1918 and 2019 to prognosticate the immensity of the health risks when a new virus outbreak occurs.
1.1. The Landscape of Pandemics Since The 20th Century
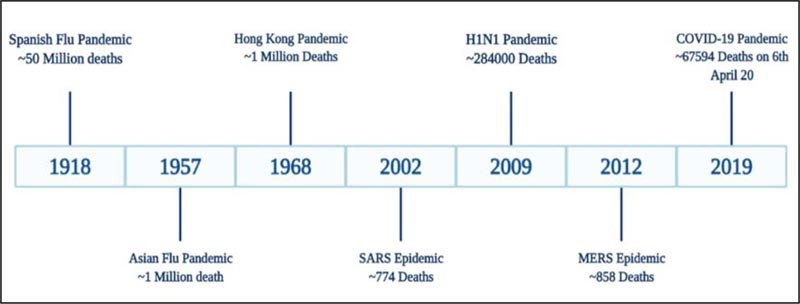
The globe was stunned following the emergence of the Spanish Flu pandemic in the year 1918, leading to the worldwide crisis. A novel, extremely pathogenic and highly transmissible strain of influenza, the H1N1 virus, caused the outbreak. The strain was able to surface even remote areas of the world due to the migration of the military personnel during World War I (WWI). Another strain of Influenza A virus, H2N2, arose in February 1957 in Singapore and the infection traveled to other regions of the globe by the summer of the same year. The outbreak was mild compared to the previous Spanish flu and was named Asian flu pandemic [7]. Similar to H1N1 infection, the outbreak appeared in different phases of the waves but was less transmissible. It caused approximately 1.1 M deaths worldwide from the year 1957-1958 [8, 9]. A decade after the outbreak of H2N2, due to genetic reassortment, H3N2 virus evolved in 1968, leading to the Hong Kong flu pandemic. Even though the virus was highly transmissible, the clinical symptoms of H3N2 infection were milder than the H2N2 [10]. The first wave of the Hong Kong flu pandemic was a major burden in the United States and Canada, while the second wave surfaced the European and Asian regions accounting for approximately 2 million deaths globally [11]. In 2009, another strain of the Influenza species, the pH1N1/09 virus, emerged that went through multiple reassortments between different influenza lineages, of which one was the 1918 human virus [12]. The other names of this virus are swine flu, Mexican flu [as it originated in Mexico] or new flu, and it caused nearly two lac deaths [13]. Besides these outbursts instigated by the influenza species, the coronaviruses, in addition, are responsible for outbreaks such as the Severe Acute Respiratory Syndrome (SARS) 2002 epidemic, the Middle East Respiratory Syndrome (MERS) and the COVID-19 pandemic. The first two pandemics were very mild that accounted for a toll of fewer than 1000 deaths worldwide. The COVID-19 pandemic has greatly influenced the world because of a highly transmissible state [3]. The Spanish flu and COVID-19 pandemic appear to be unique; as discussed above, these viruses from different origins have caused major negative impacts on the world. Visualizing the current picture of COVID-19 pandemic, there are chances that soon the pandemic may take the face of the 1918 outbreak, and currently, the world is not in a state to bear such load. Therefore, it has become a prerequisite to evaluate the course of Spanish flu and explore strategies from the previous experiences to contain COVID-19. Fig. (2) depicts the significant outbreaks that occurred due to the influenza and coronaviruses.
2. HISTORICAL BACKGROUND
In spring 1918, the first case of the H1N1 virus appeared in Kansas of the United States after military personnel showed symptoms of fever, cough and headaches. Initially, the disease impact was low in its first wave, but subsequently, the second wave was marked by high virulence and fatality [14]. This highly lethal wave emerged in an Army Training Camp near Boston in the US, and fall 1918 led to emergence of many infection cases and the deaths, eventually leading to the shortage of nurses in the United States. The third wave began in winter and spring of the year 1918, which steadily receded in the summer of the same year [15]. The influenza virus responsible for this outbreak originated from wild waterfowl birds, which are the main reservoir of this virus and caused the transmission to humans. The virus strain infectious to humans is a transformation of the avian virus strains through a phenomenon called reassortment [16]. This antigenic shift was responsible for the virus to escape from immunity in that era [17]. The literature demonstrates that pigs are vulnerable to both the strains [avian and human]. Therefore the pig is thought to be an intermediate host in the transmission process from birds to humans [18]. The H1N1 infection hustled across the globe during the WWI as the military troops of different countries served as the carriers and took the virus to their respective countries. The name of the disease outbreak arose as “Spanish Flu Pandemic” due to high mortality and morbidity, and Spain was the first to acknowledge the disease outbreak [19].
After a century passed, the new virus SARS-CoV-2 evoked and caused a pandemic. In December 2019, in Wuhan, China, a series of unexplained pneumonia cases were reported [6]. Realizing the aggressive transmission behaviour of the disease, the World Health Organization [WHO] on 31st January 2020 declared this epidemic as a Public Health Emergency of International Concern [PHEIC]. As the virus responsible for thousands of death is genetically similar to the virus which caused SARS in 2002, the International Committee on Taxonomy of Viruses [ICTV] announced the name of the virus as “severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2].” Whereas on the same day, the WHO declared the disease as ‘COVID-19’ [20]. The epidemic soon evolved into a pandemic after the disease abruptly spread to the remote areas of the world. Some studies observed the sequence homology between the viruses responsible for the 2002 [SARS-CoV] and 2019 [SARS-CoV-2] pandemic and 79.5% of sequence homology was shown [21, 22]. The SARS-CoV-2 virus is a zoonotic agent originating from animals [similar to the SARS-CoV virus] and causing infection in humans. The virus has been considered to be transmitted to humans from bats, but the intermediate host or the reservoir is still mysterious [23, 24].
3. EPIDEMIOLOGY
The 1918 flu pandemic, which emerged from the United States, was responsible for 45-50 million deaths across the globe, which accounts for 2-3% total population, including 675,000 Americans [15]. However, the exact number of deaths due to the H1N1 virus remains a sphinx-like enigma as different authors have reported diverse figures viz. 32 million [1991], 50 million [2005], 17 million [2018] [14, 25, 26].The high number of deaths were observed in the second and third wave of the pandemic. One of the striking features of this virus was the “age-specific” mortality rate, that is, it involved the young adult age group population. Conventionally, the graph of other strains of influenza viruses follows a “U-shape” curve affecting the age groups <20 and >40 years, whereas the H1N1 strain exhibited “W-shape” curve with an extra peak of mortality in 20-40 year age groups [27, 28]. One of the theories supporting such age-specific deaths explains that this behaviour may be due to the secondary pneumococcal infections to the young adults and the high virulence and pathogenicity of the virus [29-31]. Several thousands of deaths were the result of secondary bacterial infections in these positive flu patients, and the bacteria were mainly “Streptococcus pneumonia, Streptococcus pyogenes, and Staphylococcus aureus” [32]. The age group of more than 65 years accounted for only 0.6% of influenza cases. Another theory in favor of the same proposes that the influenza virus A H1N1 possesses antigenically shifted HA gene, which is novel and the population lacks immunity to it [18]. The low susceptibility of the older age group may be due to the already existing antibody against the H3 influenza A virus [the virus responsible for the 1889-91 epidemic] protected them to a certain extent [33, 34]. However, even today, the other novel segments of genes of the virus are unclear.
The COVID-19 disease emerged in December 2019 in China, and within one month on 22nd January 2020, 571 positive coronavirus cases were reported in the 25 cities of China, accompanied by a death toll of 17 patients [35]. The disease spread abruptly in Chinese provinces and other countries, resulting in 7824 confirmed positive cases of SARS-CoV-2 by 30th January 2020 [36]. According to the WHO report of COVID-19 as of 8th May 2020, the number of confirmed positive cases across the globe is ~3.7 million and ~2.3 lac deaths. The USA has become the epicentre of the disease outbreak, with ~1.2 million confirmed SARS-CoV-2 cases and ~67146 deaths. The situation in Europe is also at its peak, mainly including Spain, Italy, United Kingdom, Russian Federation, Germany, France and Turkey [37].
4. CLINICAL FEATURES
The first wave of the H1N1 pandemic was mild as compared to the second and third waves; the symptoms presented by infected patients were mild, like nasopharyngitis, cough and sore throat accompanied with systemic manifestations like fever and muscle pain [38]. Epistaxis [bleeding from the nose] was the common symptom in both mild and severe phases of the disease [39]. The symptoms in mild cases lasted no longer than 3-4 days, with the body temperature at ~1040 C [14]. Prostration was also one of the complaints of some patients, which involved a sudden onset and was of severe type [40]. The acute form of Spanish flu was found to be associated with respiratory distress and signs of cyanosis, breathlessness and progressive pulmonary oedema [41]. In extremely severe conditions where death was inevitable, the clinicians observed a deep purple colour in the patients before death. Hence the cyanosis related to Spanish flu was known as “heliotrope cyanosis” which resembles the “‘heliotrope flower” [41]. The signs of cyanosis started first appearing in the lip and ear regions and then progressed to involve other areas of the face [41]. As discussed earlier, the increased deaths were due to secondary infection from bacteria causing pneumonia and acute respiratory distress syndrome [ARDS], aggravating the illness [32]. The severe form of illness lasted up to 8-10 days, while death due to the infections mostly occurred 14-15 days after the onset of symptoms [39, 41]. Nevertheless, the clinical and radiographical presentation of the pneumonia was different from the classic case of lobar pneumonia. A clinician described the virus-infected lung superimposed with bronchopneumonia as the “lungs of the drowned” [42]. Bronchitis, pulmonary oedema and haemorrhage, and hyalinization of the alveolar tissues marked the features of lower respiratory tract infection by the H1N1 virus, which was lethal [43].
SARS-CoV-2 infection in patients exhibits a broad spectrum of features, which range from asymptomatic conditions to severe conditions like respiratory failure, sepsis, Multiple Organ Dysfunction Syndrome [MODS] [44]. Depending upon the pathogenicity of the virus and the host response, the disease can be classified as “mild, moderate, severe and critical”, as elaborated below [20, 21, 45, 46]. The incubation period of the disease ranges from 5-14 days. The majority of the patients report complaints of fever, dry cough, sore throat, headache, fatigue and diarrhoea [47].
Mild/Asymptomatic Disease: The symptoms include a mild form of pneumonia, and features of upper respiratory tract infection like sore throat, dry cough, headache, fatigue and myalgia. More than 80% of disease cases report mild conditions. Breathlessness and radiographic features are mostly absent in such conditions. Most of the patients improve from the condition and do not require any special medical attention.
Moderate Disease: This is a transient phase of the disease from mild to severe, and many times, it is of short duration as the severity of the illness increases rapidly. In addition to gentle features, the patient experiences shortness of breath and tachypnea.
Severe Disease: The severity of the condition is characterized by respiratory distress, severe breathlessness, and a respiratory rate of more than 30 breaths per minute. Further, SpO2 ≤ 93%, PaO2/FiO2 ratio < 300, and/or lung infiltrates > 50% within 24 to 48 hours may be observed; this occurs in 14% of cases. The severity is usually associated with the presence of underlying diseases like hypertension, diabetes, chronic conditions like Chronic Obstructive Pulmonary Diseases [COPD], heart disease, cancer, etc.
Critical Disease: The critical phase involving Acute Respiratory Distress Syndrome, Sepsis, MODS occurs in merely 5-6% of cases [48]. The key features of H1N1 and SARS-CoV2 are summarized in Table 1.
| Virus | SARS-CoV-2 | H1N1 |
| Estimated Reproduction Number | 2.24-3.58 (early phase) (40) | 1.7-2.o (1st wave) (41) |
| Origin | Avian (Bat) | Avian (Waterfowl) |
| Intermediate Host | Unknown | Swine |
| Transmission | Respiratory Droplets | Respiratory Droplets |
| Target Organ | Lungs | Lungs |
| Complications | Respiratory failure, MODS, Septic shock | Bronchopneumonia, Pulmonary haemorrhage |
5. DISCUSSION
The H1N1 virus and SARS-CoV-2 accountable for the death-dealing pandemics of 1918 and 2019 respectively differ in their genomic organization and pathogenicity. Despite this fact, these viruses share a similar epidemic orbit. The aim of the current review was to correlate the pattern of both outbreaks as well as explore strategies to contain the COVID-19 outbreak according to similar strategies that were utilised to halt the Spanish Flu pandemic. The Spanish Flu outbreak was the most shattering in history to date. The outbreak manifested four years after the commencement of World War I when the US military troops traveled across the Atlantic, contributing to the spread of virus around the globe [49].As discussed earlier, the 1918 outbreak had three phases recognized as the three waves of the pandemic. Based on census data, during the summer of 1918, a sharp surge in influenza and pneumonia mortality in April was observed. The fatality stats from cities in the United States suggest the visibility of the first wave surfacing in the summer of 1918. The first wave, also known as the spring wave, was well-thought-out to be mild, which was followed by a massive second wave after six months in autumn 1918 [50, 51]. This is because the infected cases during the first wave presented mild to moderate clinical features and a low death rate as compared to the second and third waves. The first wave was equivalent to the seasonal outbreaks of influenza. Estimates reveal that the virus strain which mushroomed around the world underwent mutation [4, 52]. The second wave, which instigated form France, Sierra Leone and United States, caused havoc in other states/territories like North, Central and South America, western and south Africa, Europe including Poland and Russia, northern Asia and southeast Asia, including India and China by the end of November 1918 (Fig. 3) [53]. The third wave of Influenza was undetectable in some locations. It arrived during the winter of 1918–1919 [54]. As these three pandemic waves are distinct to the Spanish Flu, we have tried to correlate the waves with the stages of COVID-19 outburst. The number of SARS-CoV2 positive patients increased from 282 on 22nd January 2020 to 7818 people on 30th January, including China and other countries following the Covid-19 situation report of WHO. According to the WHO report on 18th April 2020, there
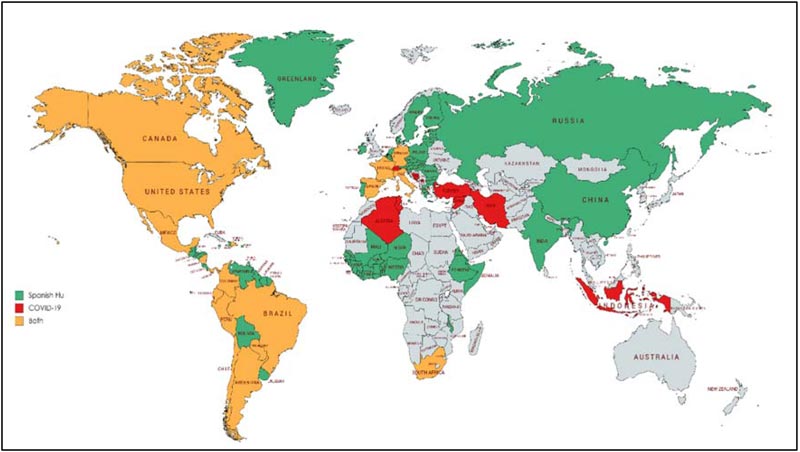
has been an exponential rise in the SARS-CoV2 infected cases from the third week of March 2020. This suggests that the second wave of COVID-19 surfaced the globe circa three months after the outbreak initiated. The early phase R0 of COVID-19 has also been found to be higher than the Spanish flu (Table 1). The community transmission stage of COVID-19 corresponds to the second wave of the Spanish Flu. Many states/territories are experiencing different stages of severity in the world, out of which the US, Italy, Spain and France account for the highest deaths (Fig. 3) [55, 56].
Spanish Influenza virus and SARS-CoV2 are novel viruses owing to their new strain and lack of immunity in humans against them. The mode of transmission and the target tissue [lungs] to cause morbidity in hominids are identical for both the viruses, though the severity of clinical manifestation may differ. Both these viruses have been proved to be fatal in cases with pre-existing chronic diseases either lung diseases, cardiac diseases, diabetes mellitus, cancer, etc [57, 58].
These viruses have grave economic consequences on a global level, disrupting the demand and supply chains. The shortage of healthcare products has also led to a panic situation. There is lasting negative psychological influence on society due to the pandemic.
CONCLUSION
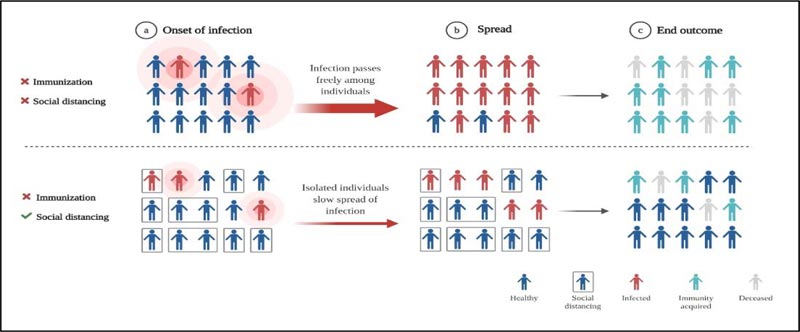
The disastrous spread of Spanish Influenza was predominantly credited to overcrowding or mass gathering [59]. Even though in the present review, we have proposed that the stage of community transmission of COVID-19 can correlate to the second wave, we have not yet faced the human toll as caused by the 1918 Spanish flu pandemic. Sanitization and disinfection strategies were followed to contain the virus spread and no vaccines or pharmaceuticals were available, and as a result, influenza ended taking the lives of millions; however, the population which was saved may have developed immunity against it [4, 51]. This herd immunity comes into effect after involving more than 50% of the population as a result of acquired resistance among the recovered patients [60, 61]. For this phenomenon to happen, time is a limited resource considering such a vast population is affected. The role of “social distancing” can add up to the effect of herd immunity and may prevent transmission of infection. Yet no vaccines or specific drugs against novel coronavirus are available (Fig. 4). Epidemics put a lot of stress on the underwhelming health care system. It will take years to overcome economic, political disruptions caused by the viruses. Therefore, the only way to prevent the coronavirus from surfacing the whole globe is “social distancing” and proper sanitization. The post-COVID-19 world will see a shift in government and private institutes. The parameters for the survival of human species will evolve and COVID-19 pandemic will become a foundation for upcoming generations in building and designing methods to resist these unknown microbes.
CONSENT FOR PUBLICATION
Not applicable
FUNDING
The authors received no funding for this article.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest associated with the manuscript.
ACKNOWLEDGEMENTS
This paper would not have been possible without the exceptional support of my institute Datta Meghe Institute of Medical Sciences. I am also grateful for the insightful comments offered by the anonymous peer reviewers at The Open Public Health Journal. The figures in the manuscript were prepared using “Biorender” software.


