All published articles of this journal are available on ScienceDirect.
A Causal not Casual Approach to Coronavirus Disease 19. Tracing the Roots of Novel Virus
Abstract
Coronavirus disease (COVID-19) has generated extraordinary circumstances worldwide like never before; India is already reeling under the health issues caused by this disease. At this critical juncture, having insights into pathogenesis is important so that unwanted panic and uncertainty regarding causative mechanisms can be curtailed. The causative pathogen of COVID-19 has been identified to be SARS-CoV-2 or also known as novel Coronavirus (nCov), which is a variant of the Coronaviruses (CoV). Through this review, we intend to present phylogenetic analysis of nCoV, epidemiology and pathogenesis of COVID-19. On the basis of nucleotide sequencing, nCoV isolates from China and US were found to have the highest similarity index of about 88.2% with two “Bat-SARS-like CoV (Bat-SL-CoVZC45 and Bat-SL-CoVZXC21. Researchers think that bat might have initiated the outbreak and an unknown wild animal might have acted as an intermediate host prior to the transmission to humans. Nasal cavity is considered to be the entry point for nCoV. Initially, a defined RBD of nCoV will locate the ACE2 receptors of Type II Pneumocytes in the alveoli, and will attach and fuse together to form a receptor host membrane. This critical step is responsible for the susceptibility of the host. Blessing in disguise is that the mutation rate of “nCoV” is much slower than “SARS CoV” and “MERS CoV”. Thus, vaccines and antiviral agents developed will not be rendered ineffective early due to slow genetic drift. The live animal markets act as highly potential centres for spill over of viruses from their reservoirs to other species and in turn humans. Such markets need to be dealt with diligently in the wake of the high risk they pose for such outbreaks.
1. INTRODUCTION
Coronavirus disease (COVID-19) has generated extraordinary circumstances like never before, world is already reeling under the health issues caused by this disease. This global pandemic has engulfed more than 200 countries and the numbers are still soaring. Reasons for resurgence and transmissibility of coronavirus are varied. At this critical juncture, having insights into pathogenesis is important so that unwanted panic and uncertainty regarding causative mechanisms can be curtailed.
The causative pathogen of COVID-19 has been identified to be SARS-CoV-2, also known as novel Coronavirus (nCov), which is a variant of the Coronaviruses (CoV). The genetic structure of CoV places it in Coronaviridae family with single strands of RNA genome. Corona in Latin means crown and its name is derived due to its spike projections from the virus membrane, which are known to be largest amongst RNA viruses’ genome [1].
The Coronavirus primarily affects the respiratory system of humans. The first instance of COVID 19 dates back to December 2019 when a few patients with a history of pneumonia were reported. This was the first emergent situation pertaining to nCoV of this decade, which originated from the animal wholesale market in Wuhan, China [2]. Origin of COVID 19 has been marred with various conjectures. It is still not clear whether a superspreading event occurred in this market or it was the reason of the initial outbreak.
All coronaviruses have a similar structural genomic expression, which consists of sixteen non-structural proteins followed by four structural proteins viz. Spike (S), Envelope (E), Membrane (M), and Nucleocapsid (N) (Fig. 1). Human Coronavirus (HCoV) are amongst the variants of Cov, which are primarily known to infect humans. These HCoVs are six in number, namely, “HCoV-NL63, Severe Acute Respiratory Syndrome coronavirus (SARS-CoV), HCoV 229E, Middle East Respiratory Syndrome Coronavirus (MERS-CoV), HCoV OC43 and HCoV-HKU1” [3]. Phylogenetically CoV has four generations, namely “Alpha-CoV, Beta-CoV, Gamma-CoV and Delta-CoV” [4]. Alpha and Beta forms of CoV infect only
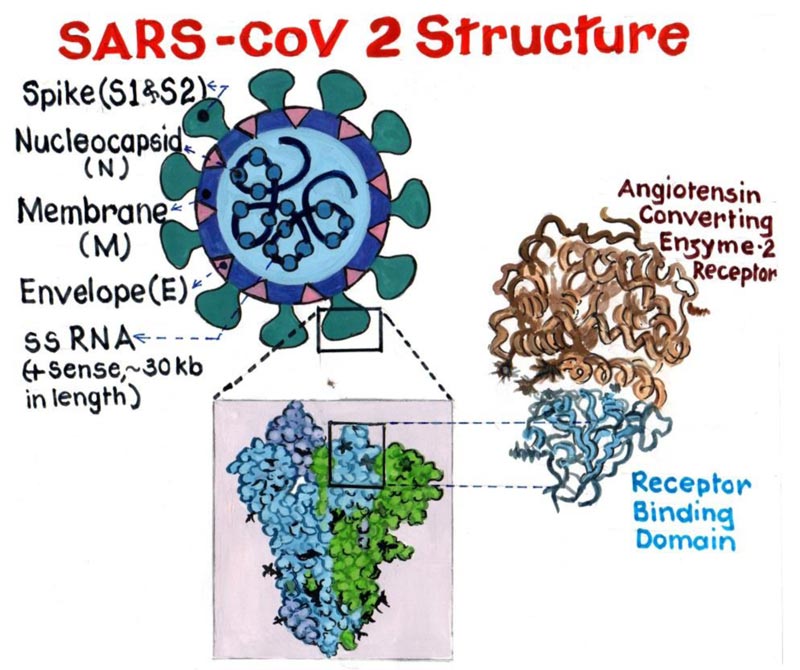
mammals and are certainly responsible for human respiratory infections and enteric disease in animals. The two major zoonotic pathogenic CoV “SARS-CoV and MERS-CoV” belong to the genus “Beta- CoV”.
The scientific interest in CoV grew after the breakout of SARS in 2002-2003. We intend to present phylogenetic analysis of nCoV, epidemiology and pathogenesis of COVID-19.
2. PHYLOGENETIC ANALYSIS
Human origins could be ascertained due to steps undertaken in knowing the human mitochondrial DNA. It helped us in getting information about movements on the planet before documentation was in place. Therefore, phylogenetic analysis is one of the standard practices for understanding the origin and evolution. This is the time we apply similar concept of phylogeny in understanding the evolution of nCoV.
Literature is based on the evidences related to the “Nucleotide sequencing and S gene pattern” in amino acid based index. On the basis of nucleotide sequencing, the isolates of nCoV, from China and US were found to have the highest similarity index of about 88.2% with two, “Bat-SARS-like CoV (Bat-SL-CoVZC45 and Bat-SL-CoVZXC21)” [5], followed by SARS Cov, which is 70.8% to 74.7%. The similarity descended to about 40.8% to 41.5% with “Canine Respiratory CoV (CRCov)” and “Bovine CoV (BCov)” of the subgenus “Embecovirus”. The “S gene” pattern similarity, for nCoV isolates with Bat SARS-like CoVs varied from 81.2% to 81.8% and for other SARS like CoVs from, 77.0% to 78.1%, respectively [6]. The similarity of nCoV with isolates of HCoV-OC43 was also found to be lower (28.0%). These distinctiveness and variabilities are indicative of a diversified nature of nCov.
The motifs of all nCoV have well modified Receptor binding domain (RBD) and the polybasic cleavage site is considered to be derived from a common predecessor. The nCoV shows divergence based on the phylogeny, despite the similarity to SARS-CoV.
The divergence is attributed mainly to the “S” protein of nCoV, which is particularly distinct from other viruses under the same subgenus Sarbecovirus. The length of the “S” protein is more than the other viruses, and at the N-Terminal region, three short insertions have been detected. Also, in the Receptor binding domain (RBD) of “S” protein four changes are found.
But, the Binding affinity of nCov RBD to Angiotensin Converting Enzyme 2 (ACE2) is found to be significantly higher than the SARS CoV RBD. This serves as the major cause for its extensive transmission from human to human [7].
The contribution of bats for spreading COVID-19 cannot be overlooked but substantial facts regarding the role of an intermediate animal between bats and humans cannot be neglected. In December 2019, when the first outbreak was reported, most bat species were in hibernating phase. Secondly, no bats were for sale in the market. In previous human viruses “SARS-CoV” and “MERS-CoV” another animal served as an intermediate host. The evolutionary relationship between CoV and bats was proposed by some researchers. It was found that the predecessors of SARS-CoV first spread through the bats of the Hipposideridae family, then Rhinolophidae family, then masked palmcivets and eventually humans [8]. With this information, available researchers think that bat might have initiated the outbreak and an unknown wild animal might have acted as an intermediate host prior to the transmission to humans [9].
While, tracing the roots of CoV it was found that Himalayan Civet was the natural host of SARS CoV. Later on, in 2005 an HKU variant in horseshoe bats was identified in Hong Kong. Ever since bats were regarded as the potential reservoir and natural host of SARS CoV [10].
Virus sampled from Rhinolopus affinis bat resembles 96% to SARS CoV 2 but divergence in its receptor binding domain impairs its precise binding to ACE 2 receptors. Malayan pangolins coronaviruses exhibit stronger kinship to SARS CoV 2 through six key receptor binding domains [9, 11].
3. PATHOGENESIS AND EPIDEMIOLOGY OF NCOV
Symptoms caused by nCoV have distinct similarities to those caused by SARS-CoV. There were 8000 confirmed cases and 800 deaths of SARS CoV in 2002-2003. It re-emerged in 2003-2004 but was in milder form, having no human to human transmission [12, 13].
Similar to “SARS-CoV” and “MERS-CoV”, “nCoV” is known to cause respiratory infections in the form of fever, cough, dyspnoea and more severe sequelae like pneumonia, ultimately progressing to Acute Respiratory Distress Syndrome (ARDS) [14]. It is interesting to know the path of the entrance of nCoV in the host cell (Fig. 2). The Spike (S) Protein, in the RBD, which is envelope-anchored facilitates the entry in cells of the host. Nasal cavity is the entry point for nCoV where it binds to epithelial cells and starts multiplying. Innate immune response is restricted at this point of time. Over the course of next few days, it travels down the respiratory tract, during which the clinical features become evident [15]. Further through the trachea-bronchial tree, it reaches up to the alveoli. Initially, a defined RBD of nCoV, will locate the ACE2 receptors of Type II Pneumocytes in the alveoli, and will attach and fuse together to form receptor host membrane. This critical step is responsible for the susceptibility of the host. Once the single stranded genome virus is engulfed, it will lead to series of pathogenic changes. Firstly, it will release the single stranded RNA into the cytoplasm. This RNA uses the host cell ribosome and undergoes translation after which this RNA gets converted into specific polyprotein molecules. Secondly, RNA dependent RNA polymerase will replicate the RNA. Simultaneously, these polyprotein molecules produced are transformed into various viral components by enzyme proteinase. These viral components get combined with the replicated RNA’s to produce a number of virus particles.
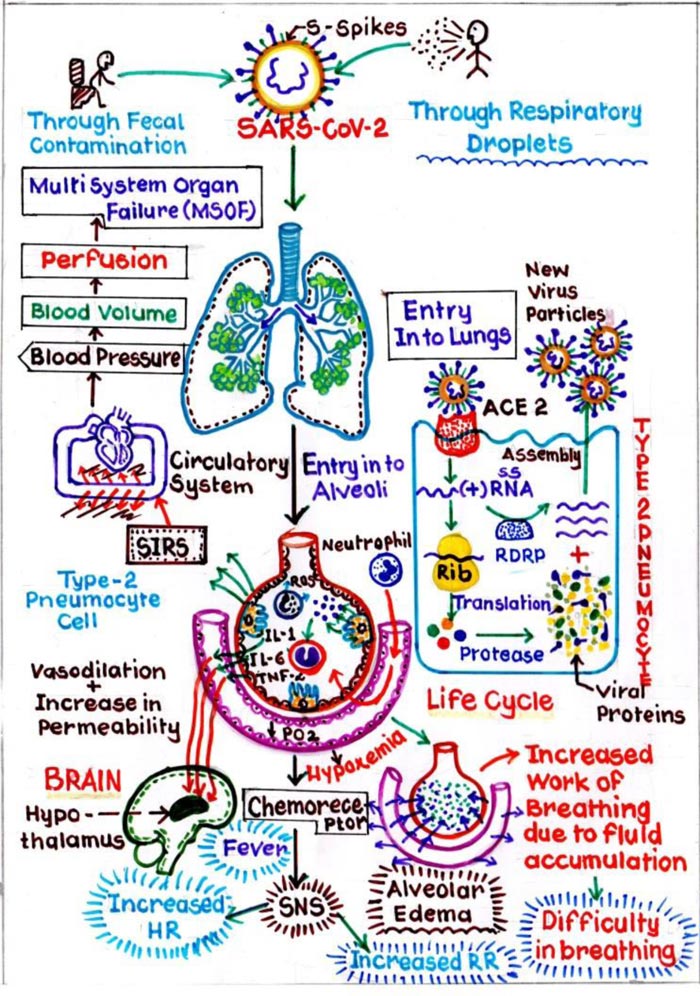
In this process of formation of viral components, Type II pneumocytes which are responsible for producing pulmonary surfactant, are destroyed. This again leads to a series of events in which initially inflammatory mediators are released. These mediators will stimulate the Macrophages, which will release Cytokine i.e. IL1, IL6 and TNFα, further causing vasodilation and increases the capillary permeability. Due to this interstitial oedema increases around the alveoli, resulting in its compression. Also, some amount of interstitial fluid enters the alveoli, resulting in alveolar oedema, and damaging the surfactant, further leading to alveolar collapse. This collapse will cause Hypoxemia, and increases the work of breathing. Secondly, during the process of release of inflammatory mediators, Neutrophils released as a first line defence to destroy the viruses will in turn land up, collapsing the alveolus itself. The process starts by the release of Neutrophils, which further stimulate the proteases and reactive oxygen species, which in turn damage the Type I pneumocytes, which has a pivotal role in gaseous exchange. Thus, the cumulative effect leads to diffuse alveolar damage, further promulgating into, acute respiratory distress syndrome (ARDS). Healing in response to this will cause more scarring and fibrosis [16].
Even asymptomatic individuals of nCov have contributed to its spread. In terms of incubation, nCov follows the trends of other human coronaviruses. The nCov can even be detected in faeces but whether it has a role in faeco-oral transmission is still to be ascertained [17]. Previously known human coronaviruses are well adapted to humans. For this to happen, adaptive mutations have to be present, which counteract host restriction factors.
Researchers discovered a couple of viral hotspots on ACE2 receptors of Type II pneumocytes of human around which naturally occurring RBM mutations occurred. This played a pivotal role in decoding the host range. The “nCoV” has a distinct capability of identifying homologous gene sequences of ACE2 receptors from a diverse pool of species, except in the species of mouse and rat. This distinct feature for partial viral adaptations stimulated viral replications and promoted the interspecies transmission [18].
COVID-19 infected individuals have demonstrated higher levels of C-reactive protein (CRP), Erythrocyte sedimentation rate (ESR), D-Dimer. Although the normal CRP is in the range of 0 mg/L to 10 mg/L, but in COVID19 infected individuals, it reaches the tune of 16.16 mg/L [14]. Also, significantly elevated blood levels of Inflammatory mediators such as Cytokines and Chemokines are also noted. Mostly Interleukin (IL1-β,1 RA,7, 8, 9, and 10), Fibroblast Growth factors (FGF- 2), Granulocyte Colony Stimulating Factors (GCSF), Granulocyte Macrophage Colony Stimulating Factor (GMCSF), Interferon Gamma (IFNγ), Interferon Gamma induced Protein (IP10), Monocyte Chemoattractant Protein 1 (MCP1), Macrophage Inflammatory Proteins (MIP1α and MIP1β), Platelet derived Growth Factor subunit B (PDGFB), Tumor Necrosis Factor (TNFα), and Vascular Endothelial Growth Factor (VEGF A) [19]. Non-invasive ventilation (NIV) is an emerging tool in treating the patient with Acute hypoxic respiratory failure (AHRF) and can prevent patients landing up in ARDS [20]. However, patients with severe hypoxia may require assisted ventilation. During the process of intubation, it is a known fact that stimulation of pharyngeal and laryngeal receptors may stimulate the tachycardia and hypertension [21], ultimately culminating in ARDS.
The mutation of CoV, in general, is on a higher aspect, when compared to other single stranded RNA viruses. This mutation rate is attributed to a lack of proof reading exoribonucleases [22]. But the blessing in disguise is that the mutation rate of “nCoV” is much slower than “SARS CoV” and “MERS CoV” [23]. Thus, vaccines and antiviral agents developed will not be rendered ineffective early due to slow genetic drift.
CONCLUSION
Coronavirus has resurfaced with the advent of “SARS-CoV-2”. COVID-19 led to unprecedented circumstances, which has put strenuous load on the healthcare services. The economy has come to a standstill. It has infused a sense of fear and uncertainty amongst people. This virus has made us understand the magnanimity of zoonotic diseases and extent to which they can go in putting the human race at risk on all fronts. The live animal markets can act as highly potential centers for spillover of viruses from their reservoirs to other species and in turn humans. Such markets need to be dealt with diligently in the wake of the high risk they pose for such outbreaks.
CONSENT FOR PUBLICATION
Not applicable
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.


