All published articles of this journal are available on ScienceDirect.
Security of Magnetic Resonance Medical Images using Region-based Lossless Image Compression in Healthcare Information Systems
Abstract
Purpose
The purpose of region-based medical image compression is to optimize the compression process by focusing on specific regions of interest within medical images. Unlike traditional compression methods that treat the entire image uniformly, region-based compression techniques identify and prioritize certain areas or regions within the image that are deemed more diagnostically significant or relevant. By allocating more resources to compressing these critical regions while reducing compression in less important areas, region-based compression methods aim to achieve higher compression efficiency while preserving diagnostic quality. This approach is particularly valuable in medical imaging, where accurate representation of anatomical structures or pathological findings is paramount for clinical diagnosis and decision-making. Region-based compression can help reduce storage requirements, transmission bandwidth, and processing time without compromising the diagnostic integrity of medical images, thereby facilitating more efficient healthcare delivery and telemedicine applications.
Methods
In this study, we utilized distortion-limiting compression techniques to optimize the compression process for specific regions within medical images. We employed lossless scalable RBC (Region-Based Compression) using Discrete Wavelet Transform (DWT) for Digital Imaging and Communication in Medicine (DICOM) images. The initial step involved medical image pre-processing, followed by segmentation to separate the image into regions of interest (ROI) and non-ROI. Compression techniques were then applied to reduce network bandwidth and storage requirements. Fractal lossy compression was employed for the non-ROI portion, while context-tree weighting lossless compression was proposed for the ROI portion, effectively compressing the image while rejecting noisy background elements. During decompression, the original medical image can be reconstructed using the reverse process. This approach optimizes storage and transmission efficiency while preserving diagnostic integrity in medical imaging applications.
Results
The experiment involved testing various medical images, and the proposed method outperformed previous techniques in terms of results. According to the findings, the improvement in Peak Signal-to-Noise Ratio (PSNR) over current techniques reached up to 24.23 dB compared to the Joint Photographic Experts Group (JPEG). Additionally, it achieved up to 12.22 dB improvement compared to other transform approaches. These significant enhancements prompted the development of a web and mobile platform for compressing and sending medical images, particularly microscopic ones, in real time.
Conclusion
This research focuses on employing wavelet transform techniques to compress the Region of Interest (ROI) within medical images. This ROI-based compression approach is particularly valuable as it retains essential diagnostic information while reducing the overall file size. Such a technique holds significant promise for telemedicine systems, especially in rural regions where network resources may be limited or constrained. By selectively compressing the most diagnostically relevant areas of medical images, this approach ensures that critical information is preserved while optimizing data transmission and storage efficiency. This can ultimately enhance access to medical imaging services and facilitate remote diagnosis and treatment in underserved areas with limited network infrastructure.
1. INTRODUCTION
In recent years, medical imaging has become a vital tool in clinical practice, enabling the observation of internal organs for diagnostic and treatment purposes. This technique significantly contributes to reducing mortality rates, decreasing hospital admissions, extending life expectancy, shortening hospital stays, and minimizing the need for exploratory surgeries. Various types of medical imaging, such as Magnetic Resonance Imaging (MRI), X-rays, Ultrasound imaging, and Computed Tomography (CT) scanning, are employed to examine the human body. The rapid development and increasing demand for these imaging modalities have led to a substantial rise in the production, transfer, and sharing of medical images [1, 2].
Telemedicine has emerged as a pivotal component of modern healthcare, offering a means to overcome geographical barriers, improve access to medical services, and facilitate timely medical interventions. Central to the efficacy of telemedicine, particularly in diagnostics and treatment planning, is the reliance on medical imaging technologies. Magnetic Resonance Imaging (MRI), with its capability to produce high-resolution images critical for accurate medical assessments, plays a significant role in this domain. However, this advancement comes with its own set of challenges, primarily associated with the management of MRI data, which is often stored in different formats. These files, characterized by their substantial size, pose significant challenges in terms of storage and transmission, especially in telemedicine scenarios where bandwidth and storage resources may be limited. The need for efficient and effective telemedicine services is not just a matter of convenience but a crucial element in ensuring equal access to healthcare, especially in remote or resource-limited settings. Efficient trans- mission of large MRI files while maintaining the integrity and quality of the images is vital for accurate diagnosis and treatment planning. Current approaches to managing these large files, such as standard compression techniques, often lead to a trade-off between file size reduction and image quality. This compromise can impede the clinical utility of the transmitted images, potentially affecting patient outcomes. Therefore, developing advanced methods to handle large medical image files without compromising their quality is essential for improving telemedicine services and ensuring better healthcare delivery [3, 4].
1.1. Problem Statement and Motivation
Medical images are stored in various formats by different imaging modalities, making retrieval, processing, and transmission challenging. The large size of medical images leads to issues with network bandwidth and storage capacity. Unlike simple text or document files, medical images contain extensive details and information, requiring more bandwidth to transmit over different types of networks. Therefore, it is crucial to reduce the volume of these images before storage or transmission, high- lighting the need for compression. Compression is defined as minimizing the size or volume of data needed to describe a given amount of information. Efficient compres- sion techniques are thus essential in telemedicine and its applications, ensuring that medical images can be effectively stored and transmitted without compromising quality.
The motivation behind this research arises from the increasing demand for efficient and effective telemedicine services, especially within the domain of medical imaging. Magnetic Resonance Imaging (MRI) is integral to diagnostics and treatment planning, providing high-reso- lution images essential for precise medical evaluations. However, the significant size of MRI files, often stored in the different formats, presents substantial challenges regarding storage and transmission. This is particularly problematic in telemedicine contexts, where bandwidth may be limited. The difficulty is further magnified in remote or resource-limited areas, where access to advanced medical imaging and prompt communication between healthcare providers and patients is vital. Addressing these challenges is essential to ensuring equitable access to healthcare and maintaining the quality and accuracy of medical assessments in telemedicine.
The primary objective of this work is to develop advanced methods for compressing and managing large medical image files, particularly MRI data, without compromising image quality. This objective aims to enhance the security and efficiency of telemedicine services by ensuring the integrity and clinical utility of medical images during transmission and storage. Furthermore, by addressing the challenges associated with the substantial size of MRI files and the limitations of current compression techniques, the proposed solutions seek to facilitate accurate diagnosis and treatment planning, thereby improving access to healthcare, especially in remote or resource-limited settings.
The primary contributions of this work are as follows:
1.1.1. Wavelet Analysis in Image Compression
Wavelet analysis is employed to approximate image information, breaking it down into sub-signals and sub-images.
1.1.2. Enhanced Pixel Approximation
The method utilizes diagonal, vertical, and horizontal features of images, along with approximating sub-signals, to improve pixel approximation for compression purposes.
1.1.3. Minimized Computational Complexity
The effective application of Wavelet Transform (WT) reduces computational complexity by providing efficient frequency and time localization.
1.1.4. Signal-based Compression
The compression process leverages WT to capture even the smallest signal features, ensuring detailed compression based on signal characteristics.
1.1.5. Optimized Lossless Compression
During transmission, the hybrid transformation method quantizes coefficients to achieve optimized, lossless compression of images.
This paper is structured as follows: Section 2 contains the literature review. Section 3 outlines the materials and methodology. Section 4 presents the analysis and findings. Finally, section 5 concludes the work.
2. RELATED WORKS
Tarun Agrawal et al. [5] discussed various state-of-the-art deep learning models, which were evaluated and compared on an unbalanced dataset for three-class brain tumor classification. The results of their experimentation indicate that the Inception models outperformed all other models for this specific classification task. Manoj Diwakar et al. [6] introduced a novel weighted function employed by fractional order total variation for CT image denoising, addressing issues such as the blocky effect. Moreover, to resolve the non-convex optimization problem for improved solutions, they examined two different methods: (i) Split Bregman and (ii) Augmented Lagrangian. These methods were evaluated using the proposed weighted fractional total variation denoising approach. Ajay Krishan Gairola et al. [7] proposed a Fully Fused Network (FFN) that incorporates an Improved Single Block (ISB) and an Improved Fusion Block (IFB) to achieve optimal performance. This approach involves the development of a convolutional neural network-based model for multi-class recognition of skin images. The ISB is utilized to segment diseases in the skin images, enhancing the overall accuracy and efficiency of the network. Sunil Kumar et al. [8] conducted a comprehensive examination of how machine learning enhances the exploration of imaging modalities in detecting prominent lung diseases. Their review encompasses the utilization of various machine learning paradigms, recent advancements in imaging modalities, and an overview of publicly available datasets used in this field. This research provides insights into the evolving landscape of medical diagnostics through advanced computational techniques. Junbo Peng et al. [9] proposed an efficient approach involving a conditional denoising diffusion probabilistic model (DDPM). They employed a time-embedded U-net architecture integrated with residual and attention blocks. This method aims to iteratively transform a white Gaussian noise sample into the desired CT distribution conditioned on CBCT (cone-beam computed tomography). This innovative technique showcases advancements in utilizing deep learning for enhancing imaging modalities through sophisticated probabilistic modeling. Kiran et al. [10] introduced a Singular Value Decomposition (SVD)-based method for compressing medical images. This technique leverages SVD to decompose the image matrix into singular vectors and values, thereby reducing redundancy and achieving compression while preserving essential diagnostic information. SVD-based compression methods are known for their effectiveness in medical imaging due to their ability to retain image quality and facilitate efficient storage and transmission of medical data. Bharath K N et al. [11] elucidated optimal machine learning-based techniques for medical image compression in smart healthcare applications. Their research focuses on leveraging advanced machine learning models to achieve efficient compression while maintaining diagnostic quality. This approach aims to enhance the storage, transmission, and retrieval of medical images, thereby supporting improved healthcare delivery through smart technology integration.
3D medical images often exhibit excellent compression performance, and decompressed 3-D image quality is a crucial component. In addition, because of this, the preferred techniques for transmitting the decompressed and compressed medical images are lossless or bit-preserving algorithms [12-14]. Since quality and compres- sion performance are mutually exclusive, compression techniques without loss have high-quality images with minimal performance of compression [15, 16]. Further- more, because of this unusual inverse relationship, one of the two performance metrics must be sacrificed: image quality or compression efficiency. Therefore, the best image compression technique is one that equally enhances both aspects [17, 18]. The latest models of compression use computationally intensive deep learning-based approaches [19].
Applied 2D orthogonal WT coefficients and the local estimated noise were used to code and quantify the modified image sensitivity. A significant amount of compression is provided by the human visual system [20]. Big medical data CS, fractional Fourier and chaos in image encryption, fusion, and compression Transforms were applied concurrently. The method was created to decrease data and Keyboard simplification [21]. The development of secure and lossless digital picture watermarking to protect the privacy of patient information in databases, such as DWT and DCT databases. Performance PSNR measure- ments and their relationships to the entire image are development elements diminution [22]. The quick and secure transfer of primary images was accomplished via DWT online. The outcomes demonstrated that the algorithms convey critical images swiftly and safely [23]. The 3-D hierarchical listless clock algorithm (3D-HLCK), a modified 3-D block coding system with a listless form, was proposed by Senapati et al. [24]. The 3-D set partitioning embedded block approach (3D-SPECK), developed by Tang and Pearlman [10], encodes 3-D volumetric picture data by taking advantage of the interdependence in every dimension. An end-to-end learning-based system for 3-D volumetric picture reduction was created by Chen et al. [25]. The approach predicts the entropy coding distribution using inter and intra-slice information. Additionally, it makes use of two innovative gating strategies to enhance the aggregation of inter and intra-slice information. The lossless compression approach by Nagoor et al. [26] presented a neural network trained to be a 3-D data predictor for volumes of medical images that contain images with tones and colors of 65,536 levels. Furthermore, Zerva et al. [27] suggested a modification to the typical Wavelet Difference Reduction (WDR) method employing Mean Co-located Pixel Difference (MCPD) to determine the ideal slice number which demonstrates the greatest correlation in temporal and spatial domain. According to an increased Structural Similarity index (SSIM) and PSNR, the slices with significant spatiotemporal coherence are stored as a single volume. Images are compressed using a variety of unchangeable standards. The Joint Photographic Experts Group (JPEG) is the most utilized standard for medical purposes. Furthermore, by including compressed data detailing the differences between the compressed and original image in an associated DAC file.
Makarichev et al. [28] changed the irreversible Discrete Atomic Compression (DAC) technique. A rebuilt image without any distortions is generated by adding to the compressed image. Golomb Rice coding, along with its architecture of hardware, was used by Lee et al. [29] to design a high-throughput image compression system. For aesthetically lossless, low-latency, lightweight picture coding, Descampe et al. [30] suggested the JPEG-XS compression technique. A comparable compression ratio to that of JPEG 2000 is achieved by this worldwide standard. A hybrid strategy was developed by Min et al. [31] to compress 3-D medical photographs. A hybrid algorithm divides medical data into several sections using the anatomical properties of the medical images. The best predictors in each area are then created by a deep neural network. A Back Propagation (BP) trained neural network and a fractional order memristive chaotic circuit were used by Yang et al. [32] to build an image compression-encryption technique. A loss-less compression method based on the Multi-Layer Perceptron (MLP) neural network was developed by Rhee et al. [33]. Prediction errors and contexts are output from the MLP, and for adaptive arithmetic encoders, they are given as input. Long Short-Term Memory (LSTM) neural networks were used by Zhu et al. [34] to construct a predictor for lossless compression. Xu et al. [35] worked on enhancing the Singular Value Decomposition (SVD) technique utilizing a singular vector sparse recovering method, which is one example of recent irreversible compression techniques. An image compression framework was created by Guo et al. [36] for computer vision applications in embedded devices. The trade-off between visual performance and memory traffic is used by the framework. A compression technique for medical images was developed by Sadchenko et al. [37] using a sample decimation approach while considering the unique characteristics of medical images. ANNs are used in several lossy techniques to boost compression ratios. Convolutional neural networks (CNN) were utilized by Dua et al. [38] to compress hyperspectral pictures. The max pooling, convolution, and auto-encoder layers of CNN are combined by the method to decrease the dimension of the image and create a compressed image. Furthermore, by using the CNN decoder and transpose convolution layer to reverse the CNN's processes, the image can be reconstructed with lost information.
Existing region-based image compression methods offer valuable insights and strategies that can be leveraged to enhance the proposed wavelet-based efficient medical image compression system. Here are some potential outcomes of existing methods and how they can be integrated or improved upon in the proposed system:
2.1. Improved Compression Efficiency
Existing region-based compression methods have demonstrated success in achieving higher compression ratios while preserving important diagnostic information within specific regions of interest (ROI). Moreover, by integrating adaptive compression techniques based on image content and utilizing wavelet transforms tailored to ROI, the proposed system can further optimize compres- sion efficiency, ensuring minimal loss of relevant image details.
2.2. Preservation of Diagnostic Information
The proposed system can benefit from the outcomes of existing methods by prioritizing the preservation of diagnostic features within ROI. Furthermore, by incorporating advanced compression algorithms that prioritize the encoding of critical anatomical structures or pathological findings, the system can ensure that essential diagnostic information is accurately represented in the compressed images.
2.3. Enhanced Visual Quality
Existing region-based compression methods emphasize the importance of maintaining visual quality, particularly in areas critical for diagnosis. In addition, by employing sophisticated compression algorithms and wavelet-based reconstruction techniques optimized for preserving image fidelity within ROI, the proposed system can enhance visual quality and reduce artifacts, resulting in clearer and more detailed compressed images.
2.4. Adaptability and Customization
The proposed system can draw from existing methods' adaptability to specific applications and imaging modalities. Additionally, by offering customizable compression parameters and adaptive compression strategies tailored to different types of medical images (e.g., MRI, CT scans, ultrasound), the system can accommodate diverse imaging requirements and optimize compression performance for various clinical scenarios.
Incorporating insights and strategies from existing region-based image compression methods can help the proposed wavelet-based efficient medical image compres- sion system overcome challenges and achieve superior compression performance, ensuring high-quality com- pressed images suitable for clinical diagnosis and analysis.
3. PRELIMINARIES
3.1. Region of Interest
As per the constraints of lossy and lossless compression techniques, the fundamental notion of ROI is established. The compression ratio for well-known lossless compression techniques is roughly its original size of 25%, however, in a lossy encoder, it is considerably higher (up to 1%). Here, the data will be lost. Then, due to the loss, some visual components will be impaired, which is crucial for diagnosis, as shown in Fig. (1). Therefore, a hybrid approach that would handle the diagnostically important section (ROI) and give a high compression ratio is required. In medical applications, where some aspects of the image are more crucial for diagnosis than others, ROI functionality is crucial. The diagnostically important information is typically restricted to very tiny regions, between 5 and 10% of the entire image area, in medical imaging. These regions, in such circumstances must be highly encoded compared to background. These regions must be sent first with a high priority during image transmission for telemedicine applications.
4. MATERIALS AND METHODS
Region-Based Compression (RBC), Progressive Transmission, and Lossless Compression are fundamental features of a compression technique suitable for telemedicine applications. RBC allows users to select any arbitrary shape as the Region of Interest (ROI), providing flexibility and adaptability to various diagnostic needs. Lossless compression techniques such as Huffman, Arithmetic, Run-Length Encoding (RLE), and Lempel-Ziv-Welch (LZW), among others, are employed to compress the ROI, ensuring that no data is lost during compression. Meanwhile, the Set Partitioning in Hierarchical Trees (SPIHT) algorithm is utilized to compress the Non-ROI portion following a wavelet transform, maintaining the integrity of image details. The latest advancements in picture compression have centered around wavelet-based methods, which offer unique advantages such as the ability to work with multiple resolutions simultaneously, a feature not available in other compression approaches. This capability enables efficient handling of diverse image characteristics and resolution requirements, making wavelet-based methods highly effective for medical image compression.
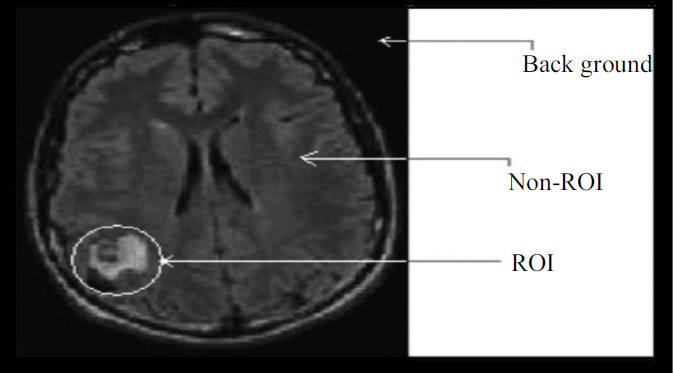
Medical image’s various parts.
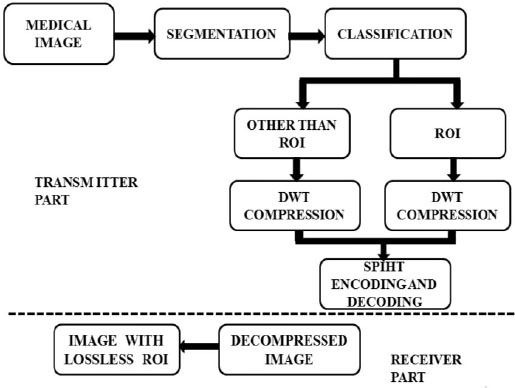
Algorithm block diagram.
In Fig. (2), the proposed block diagram outlines a region-based image compression framework tailored for medical imaging applications. The process commences with the segmentation of the input medical image into distinct regions of interest (ROI) and non-ROI components. Mathematically, this segmentation can be represented as (1):
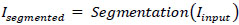 |
(1) |
Where (I_input) denotes the original medical image, and (I_segmented) represents the segmented image with identified ROI and non-ROI areas. Subsequently, the ROI portion undergoes compression using the Discrete Wavelet Transform (DWT), a technique well-suited for capturing spatial frequency information efficiently. The DWT operation on the ROI region can be expressed as Eq (2):
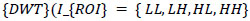 |
(2) |
Where (I_{ROI}) is the segmented region of interest, and (LL, LH, HL, HH) corresponds to the approximation and detail coefficients at different scales. These coefficients are then encoded using the Set Partitioning in Hierarchical Trees (SPIHT) algorithm, known for its ability to achieve high compression ratios while preserving image quality. The encoding process is mathematically represented as Eq (3):
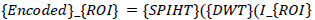 |
(3) |
The non-ROI areas, less critical for diagnosis, may undergo a simpler form of compression or be stored without compression to balance resource allocation. The final compressed image integrates both the compressed ROI and the non-ROI segments, optimizing storage and transmission within healthcare systems Eq (4):
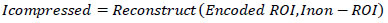 |
(4) |
This comprehensive approach, depicted in block diagram 2, underscores the systematic workflow designed to enhance healthcare information management by reducing storage demands and facilitating efficient data transmission while upholding the diagnostic integrity of Magnetic Resonance Medical Images.
MRI or CT image has mainly three parts:
I. ROI Part.
II. Non ROI part.
III. Background.
Radiologists with expert training choose the ROI. The ROI mask is created to cover the foreground completely, and background pixel values are set to zero depending on the selected area. Despite having a dark appearance, the background regions do not contain any values for the grey scale. Cross-section image of a medical condition is represented statistically as shown in Fig. (3).
Using Eq. (4), the background is set to zero:
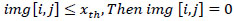 |
(5) |
The image’s threshold value (img) backdrop is indicated here by the variable x_th. Since the backdrop is not necessary, lowering its content to zero will fetch us the lossless compression completely, creating an image that is ready for processing.

Statistical representation of medical image’s cross-sectional view.
Flow diagram of the coding algorithm.
Additionally, morphological operations, which have a value of “1” in the front and “0” in the background, are used efficiently. Once the Non-ROI part and ROI (IMG_ROI) image parts have been separated, the image will be ANDed logically with the mask, as shown in Eq. (5).
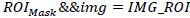 |
(6) |
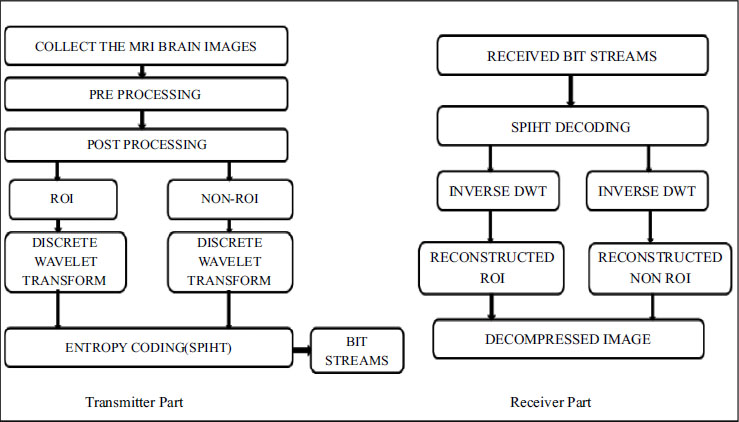
Fig. (4) illustrates the flow diagram of the proposed compression algorithm, delineating various fundamental steps involved in the process:
4.1. Image Reading and Dimension Extraction
The algorithm begins by reading the image from the database and extracting its dimensions to establish the framework for subsequent processing.
4.2. Background Removal
Background removal is performed by applying a thresholding technique, separating the foreground (ROI) from the background.
4.3. ROI Selection and Separation
The user selects the Region of Interest (ROI), and the algorithm separates the image into Non-ROI and ROI based on this selection.
4.4. Compression Level Specification
The user specifies the desired level of compression, providing flexibility to adjust the compression ratio according to specific requirements.
4.5. Wavelet Decomposition
Wavelet decomposition is applied to both the ROI and non-ROI regions, enabling the image to be represented in a multi-resolution format suitable for compression.
4.6. Wavelet Reconstruction
The algorithm performs wavelet reconstruction by recursively combining the decomposed ROI components, effectively reconstructing the compressed image.
4.7. Quality Evaluation
Finally, the reconstructed image's quality is evaluated using metrics such as Peak Signal-to-Noise Ratio (PSNR) and Mean Squared Error (MSE), comparing it to the original image to assess the effectiveness of the compression algorithm.
This flow diagram provides a comprehensive overview of the compression process, from image preprocessing to quality evaluation, offering insights into each stage of the algorithm's operation.
5. RESULTS AND DISCUSSION
In this section, we thoroughly evaluate the perfor- mance of the proposed methodology. The simulation was conducted using MATLAB 2021b, offering a robust platform for accurate analysis [39, 40]. The results derived from simulating a specific set of test images using our proposed algorithm are meticulously detailed in Table 1. Furthermore, to validate our findings rigorously, we utilized medical images sourced from the Open-i image database [41], which offers a diverse array of image data for evaluation. These images encompass a spectrum of sizes, ranging from dimensions such as 256×256 and 512x512, to 1024x1024 pixels, facilitating a thorough assessment across various resolutions. This compre- hensive approach ensures robust evaluation and validation of our algorithm's performance across different image characteristics.
 Fetal Ultrasound1 (image1) |
 Brain(image2) |
 Hand (image3) |
 Foot (image 4) |
 Ligament (image5) |
 Leg (image 6) |
 Skull1 (image 7) |
 Tumor MRI (image 8) |
 Fetus (image 9) |
 Fetal Ultrasound2 (image 10) |
 Skull2 (image 11) |
 Chest (image 12) |
5.1. Evaluation Metrics
In order to thoroughly evaluate the performance of the proposed region-based medical image compression technique, several key metrics are computed. These metrics include the Compression Ratio (CR), Structural Similarity Index (SSIM), and Peak Signal-to-Noise Ratio (PSNR). Each of these metrics plays a crucial role in assessing the effectiveness of the compression method and its impact on image quality.
5.1.1. Peak Signal-to-noise Ratio (PSNR)
Peak Signal-to-Noise Ratio (PSNR) between any two pictures is calculated in decibels (dB). The quality of the compressed and original images is evaluated by ratio. The PSNR scales from 0 to infinity; the greater the PSNR, the higher the compressed image quality. The equation for PSNR is given by Eq (7).
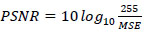 |
(7) |
Where MSE is given by Eq. (8)
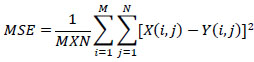 |
(8) |
5.1.2. SSIM
The visual distortion between an original and compressed image is represented by the measure known as SSIM. The SSIM is a 2-variable function between the two pictures x and y, and it is calculated between a pair of the two images' local square overlapping windows x and y. Formula (9) defines the SSIM calculation.
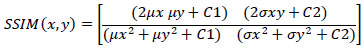 |
(9) |
5.1.3. Compression Ratio (CR)
The Compression Ratio quantifies the degree of compression achieved by comparing the size of the original uncompressed medical image to the size of the compressed image. It is calculated as the ratio of the size of the original image to the size of the compressed image. A higher CR indicates a more efficient compression process, where a larger reduction in file size has been achieved without significant loss of image information. Eq. (10) defines the CR as the bitstream ratio of original and compressed images.
 |
(10) |
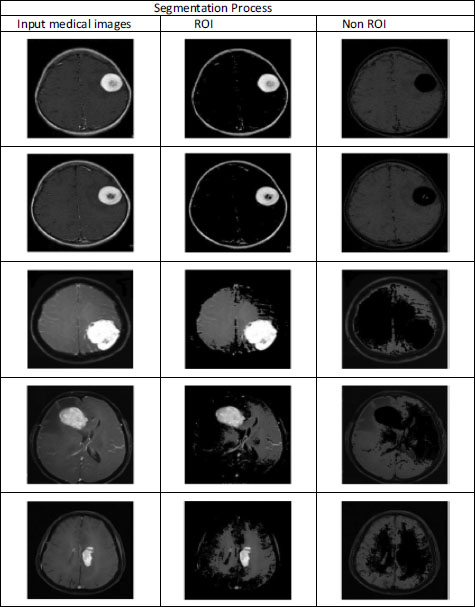
In this study, a CT scan image of a brain with tumors was selected as the primary image for analysis. The image was obtained from Radiopaedia.org, a reputable online resource for medical images. Fig. (5) illustrates the corresponding segmentation process, which involves delineating and identifying specific regions or structures within the brain image. This segmentation process is crucial for isolating areas of interest, such as tumors, from the surrounding brain tissue. Additionally, by accurately segmenting the image, researchers can focus on analyzing and processing the relevant regions, facilitating further investigation into the characteristics and properties of the tumors. This segmentation step plays a vital role in the overall assessment of the proposed region-based medical image compression technique, as it ensures that only the pertinent regions are considered during the compression process, thereby optimizing the compression results for diagnostic purposes.
In the initial stage of image processing, several pre-processing steps are applied to ensure optimal image quality and suitability for subsequent analysis:
5.1.3.1. Re-sampling of Image
The image undergoes re-sampling to standardize its resolution or size, ensuring consistency across different images and facilitating uniform processing.
5.1.3.2. Enhancement of Grey Scale Contrast
Techniques are employed to enhance the contrast of the image in grayscale, improving the visibility of details and structures within the image.
5.1.3.3. Removal of Noise
Various noise reduction methods are implemented to eliminate unwanted artifacts or disturbances from the image, enhancing clarity and accuracy.
Segmentation process of medical images.
5.1.3.4. Mathematical Operations
Mathematical operations may be applied to manipulate the pixel values of the image, adjusting brightness, contrast, or other parameters as needed.
5.1.3.5. Manual Correction
In some cases, manual intervention may be required to correct any distortions, artifacts, or anomalies present in the image, ensuring accuracy and reliability.
5.1.3.6. RGB to Gray Scale Image Conversion
If the original image is in RGB (Red, Green, Blue) format, it is converted to grayscale to simplify processing and analysis, as grayscale images contain only intensity information without color.
Moreover, by undergoing these pre-processing steps, the medical image is optimized for subsequent analysis, ensuring that any abnormalities or features of interest are accurately captured and represented for diagnostic interpretation.
After the initial pre-processing phase, the next step involves enhancing the grayscale image. Image enhance- ment aims to optimize the digital image to ensure it is well-suited for further analysis. During this stage, various adjustments are made to the image to enhance its clarity and suitability for image analysis. One critical aspect of enhancement involves removing film artifacts, such as labels and marks, that may be present on the MRI image. Additionally, high-frequency components that could potentially interfere with analysis are also removed or minimized. Fig. (6) illustrates the different stages of preprocessing and the corresponding results, showcasing the transformation and improvement of the grayscale image throughout the enhancement process. These enhancements are essential for maximizing the quality and interpretability of the medical image, ultimately facilitating more accurate and reliable analysis for diagnostic purposes.
• Image-Enhancement Methods.
• Morphological operators are used to filter.
• Histogram equalization.
• Removal of Noise with Wiener filter.
Image segmentation is a crucial step in medical image analysis, involving the partitioning of an image into multiple regions or segments based on shared properties such as gray-level intensity, color, texture, brightness, and contrast. This process serves several important purposes:
5.1.3.6.1. Study of Anatomical Structure
Segmentation allows for the detailed examination and analysis of anatomical structures within the medical image, enabling clinicians to identify and study specific regions of interest with precision.
5.1.3.6.2. ROI Identification
By segmenting the image, regions of interest (ROIs) containing abnormalities or areas of clinical interest, such as tumors or lesions, can be accurately delineated and identified for further analysis and diagnosis.
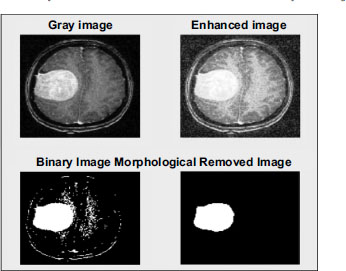
Various stages of developed software for image.
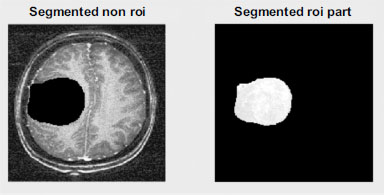
Segmentation of the ROI part of the image.
5.1.3.6.3. Measuring Tumor Growth
Segmentation facilitates the quantification of tumor growth by accurately measuring the volume of tissue occupied by the tumor. This quantitative analysis provides valuable insights into disease progression and response to treatment.
5.1.3.6.4. Treatment Planning for Radiation Therapy
Segmentation plays a crucial role in treatment planning for radiation therapy by assisting clinicians in delineating target volumes and critical structures. This information is essential for calculating the optimal dose of radiation to deliver to the tumor while minimizing damage to surrounding healthy tissues.
Overall, image segmentation is a fundamental technique in medical imaging that enables precise analysis, diagnosis, and treatment planning. In addition, by accurately delineating anatomical structures and identifying regions of interest, segmentation enhances the effectiveness of medical image analysis and contributes to improved patient care outcomes.
After the segmentation process, the next step involves the application of Discrete Wavelet Transform (DWT) to both the non-ROI and ROI parts of the image. DWT is a powerful signal processing technique that decomposes an image into its constituent frequency components, allowing for multi-resolution analysis. This decomposition is performed hierarchically, generating a set of approxi- mation and detail coefficients at different scales.
Fig. (7) illustrates the application of DWT to the segmented image. For both the Non-ROI and ROI regions, the image is decomposed into approximation and detail coefficients across multiple scales. This decomposition effectively captures the image's spatial and frequency characteristics, enabling efficient representation and compression.
Additionally, by applying DWT, the image is transformed into a format that facilitates efficient compression while preserving important image features. This transformed representation allows for effective data reduction without significant loss of diagnostic infor- mation. Additionally, DWT enables the extraction of relevant image features that can be utilized for further analysis, such as feature extraction for classification or pattern recognition tasks. Overall, the application of DWT after segmentation enhances the image processing pipeline by providing a versatile and efficient method for the representation and analysis of medical images. This step is crucial for optimizing the performance of subsequent compression and analysis techniques in medical image processing workflows.
In our approach, users have the flexibility to select the number of levels for Discrete Wavelet Transform (DWT) according to their preferences. In this study, we have opted for a 4-level transform, which offers a balance between granularity and computational efficiency. Fig. (8) illustrates the graphical representation of the DWT decomposition at different levels, showcasing the hier- archical structure of the transformed image.
Once the DWT is applied to the medical image, compression, and analysis are performed on the resulting frequency bands. Fig. (9) provides a visual representation of the DWT compression process and highlights the analysis of different frequency bands. Additionally, by decomposing the image into multiple frequency components, DWT enables the extraction of relevant information from various scales and orientations, facilitating more effective compression and analysis. Through this approach, we can effectively leverage the multi-resolution properties of DWT to capture both coarse and fine image details. This allows for more efficient compression while preserving important diagnostic information. Additionally, the analysis of different frequency bands provides valuable insights into the underlying structure and characteristics of the medical image, aiding in diagnostic interpretation and decision-making.
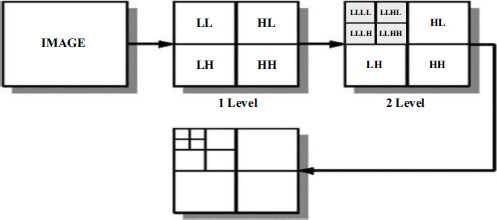
Discrete wavelet transform levels.
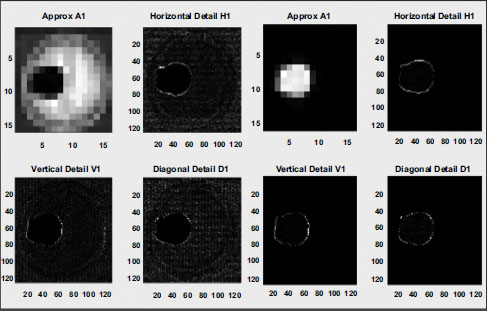
DWT compression.
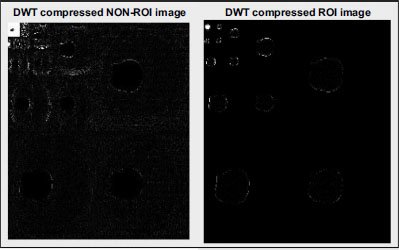
Non-ROI and DWT-compressed ROI image.
Overall, the integration of DWT into the image processing pipeline enhances the versatility and effectiveness of medical image compression and analysis techniques, contributing to improved diagnostic accuracy and efficiency in healthcare applications.
After applying Discrete Wavelet Transform (DWT) to both the non-ROI and ROI regions of the medical image, compression is performed using Set Partitioning In Hierarchical Trees (SPIHT) encoding and decoding techniques. SPIHT is an extension of the Embedded Zerotree Wavelet (EZW) algorithm, known for its efficiency in image compression.
Fig. (10) illustrates the compressed non-ROI and ROI regions obtained using DWT. These compressed regions are then encoded using the SPIHT algorithm, which significantly improves upon its predecessor by introducing innovative approaches to coefficient partitioning and refinement information transmission. One notable feature of the SPIHT bitstream is its compactness, achieved through a two-step process involving refinement pass and sorting pass. During the refinement pass, refinement information is transmitted to further improve compression efficiency. In the sorting pass, coefficients are organized into lists such as the Largest Intensity Sequences (LIS), Last Significant Pixels (LSP), and Insignificant Pixels (LIP), as depicted in Figs. (11 and 12).
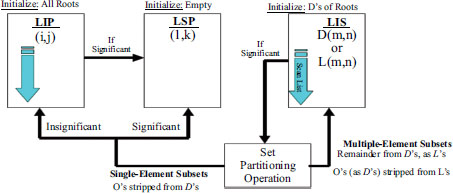
Sorting pass in SPIHT algorithm.
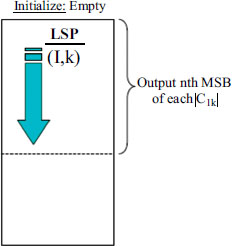
Refinement pass in SPIHT algorithm.
Similarly, by employing SPIHT encoding, the compressed image data is efficiently organized and transmitted, maximizing compression ratios while preserving essential image features. This approach ensures that diagnostic information is retained while minimizing storage requirements and transmission bandwidth, making it well-suited for medical image compression applications.
Following the compression phase, the process proceeds to decompression, which involves reconstructing the original image from the compressed data. Initially, decoding is performed using the SPIHT algorithm, which efficiently deciphers the compressed bitstream and retrieves the encoded coefficients.
The Non-ROI part is reconstructed using the inverse Discrete Wavelet Transform (DWT). This process reverses the DWT transformation applied during compression, restoring the Non-ROI region to its original state. The resulting decompressed image of the Non-ROI part is depicted in Fig. (13). Similarly, the ROI part undergoes reconstruction using the inverse DWT, as illustrated in Fig. (14). By reversing the DWT transformation applied during compression, the original details and features of the ROI region are restored, ensuring that critical diagnostic information is preserved. Through this decomposition process, the compressed image data is effectively reconstructed, allowing for accurate visualization and analysis of both the non-ROI and ROI regions. Likewise, by retaining essential image features and details, the decompressed images facilitate accurate diagnostic interpretation and decision-making in medical imaging applications.
Figs. (15 and 16) provide a detailed analysis of the compression ratio (CR) and Peak Signal-to-Noise Ratio (PSNR) for a range of MRI images, illustrating the efficacy of the proposed compression method. Our algorithm exhibits notable performance by achieving significant compression ratios without compromising the quality of the reconstructed images, as indicated by consistently high PSNR values. This graphical representation underscores the effectiveness of our approach in balancing compression efficiency with image fidelity, which is crucial for applications demanding both storage economy and diagnostic image integrity. These readings indicate a discernible level of quality in the reconstructed images, where higher Peak Signal-to-Noise Ratio (PSNR) values typically signify superior reconstruction fidelity. Beyond PSNR, Signal-to-Noise Ratio (SNR) was also assessed to gauge the extraction performance further. Evaluating both metrics provides a comprehensive assessment of our method's ability to reconstruct images accurately while maintaining fidelity and minimizing noise, which is crucial for ensuring reliable diagnostic and analytical outcomes in medical imaging applications.
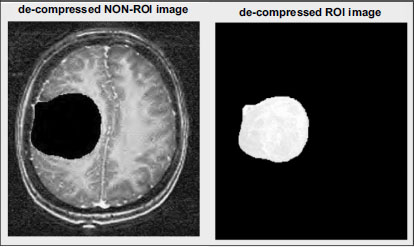
Decompressed images of ROI-based encoding with respect to level.
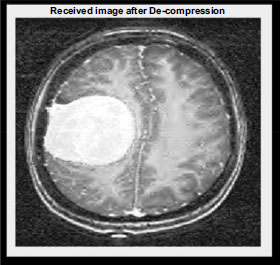
Reconstructed received medical image.
Furthermore, to provide a comprehensive evaluation, we applied the suggested algorithm at a bit rate of 0.5 bits per pixel, resulting in a compression ratio of 3. Table 2 presents the results in terms of PSNR, Structural Similarity Index (SSIM), and CR for five photos in our dataset. Notably, the PSNR values range from 80 to 88, indicating excellent image fidelity. A PSNR value exceeding 88 signifies outstanding image quality, closely resembling the original image, while values above 80 indicate very good image quality. Furthermore, the SSIM values range from 0.9 to 0.98, with values nearing 1 indicating nearly identical images. This high level of similarity corroborates the promising outcomes of our method. Notably, three out of every five photos exhibit PSNR values exceeding 85, underscoring the effectiveness of the proposed strategy in preserving image quality during compression.
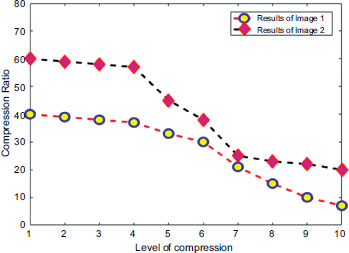
Compression ratio analysis with respect to the level of distortion.
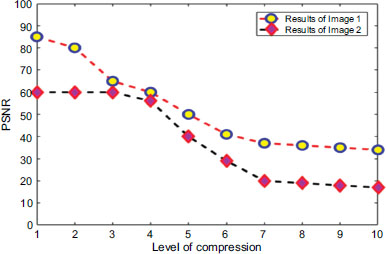
PSNR analysis with respect to the level of distortion.
| Input Medical Images | PSNR (db) | SSIM | CR (%) |
|---|---|---|---|
| Image 1 | 81.22 | 0.9493 | 3.7 |
| Image 2 | 87.45 | 0.9567 | 2.8 |
| Image 3 | 84.98 | 0.9812 | 2.6 |
| Image 4 | 86.68 | 0.9323 | 2.75 |
| Image 5 | 82.65 | 0.8321 | 2.56 |
| Image 6 | 73.23 | 0.8461 | 3.2 |
| Image 7 | 81.24 | 0.8743 | 2.7 |
| Image 8 | 89.98 | 0.8928 | 2.9 |
| Image 9 | 87.15 | 0.9010 | 2.8 |
| Image 10 | 87.09 | 0.9230 | 2.65 |
| Image 11 | 81.25 | 0.889 | 2.56 |
| Image 12 | 80.28 | 0.893 | 2.4 |
| Input Medical Images | Image Size | Size of ROI in Entire Image | % of Saving Pixel |
|---|---|---|---|
| Image 2 | 256 × 256 | 75 × 75 | 91.41 |
| Image 3 | 512 × 512 | 200 × 200 | 84.74 |
| Image 8 | 1024 × 1024 | 600 × 600 | 65.66 |
In summary, the proposed compression method achieves notable compression ratios while maintaining high-quality reconstructed images, as evidenced by the PSNR and SSIM metrics. These results demonstrate the efficacy of our approach in efficiently compressing MRI images without significant loss of image fidelity.
The computational complexity of our proposed system for compressing the Region of Interest (ROI) within the entire image is a critical aspect to consider. Our method focuses on efficiently compressing specific regions of interest, which are often smaller subsets within larger medical images. This targeted approach involves initial segmentation and identification of the ROI using advanced image processing techniques, which can vary in complexity depending on the size and complexity of the image dataset. Once the ROI is delineated, our compression algorithm applies optimized techniques such as predictive coding or wavelet transforms tailored specifically for the ROI. This approach minimizes computational overhead by concentrating compression efforts on the most diagnostically relevant parts of the image, thus optimizing both processing time and storage efficiency. Then, by strategically allocating computational resources to the ROI, our system enhances overall performance without compromising the quality or integrity of the medical image data, ensuring robust and efficient handling of diagnostic information in clinical settings. The percentage of saving pixels from the entire image is calculated in Eq. (11).
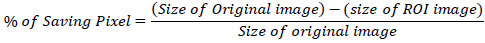 |
(11) |
Table 3 provides a summary of the computational complexity of our proposed ROI-based image compression system across different input medical images. Each row details the image size, the size of the Region of Interest (ROI) within the entire image, and the percentage of pixel savings achieved through compression.
For Image 2, which is 256x256 pixels, the ROI occupies an area of 75x75 pixels, resulting in a substantial saving of 91.41% of pixels. This suggests that the computational complexity is primarily influenced by the size of the ROI relative to the entire image. Smaller ROIs generally require less computational effort for segmentation and compression compared to larger ones. Image 3, with dimensions of 512x512 pixels and an ROI size of 200x200 pixels, achieves a pixel saving of 84.74%. Despite the larger image size and ROI, the system efficiently manages computational complexity by focusing compression efforts on the designated ROI area. In Image 8, which measures 1024x1024 pixels, the ROI expands to 600x600 pixels, resulting in a pixel savings of 65.66%. Here, the larger size of both the image and the ROI increases computational demands, particularly in the segmentation and compression stages.
Overall, the computational complexity of our ROI-based image compression system correlates closely with the size of the ROI and the percentage of pixel savings achieved. Smaller ROIs generally lead to higher compres- sion ratios and lower computational overhead, whereas larger ROIs necessitate more intensive processing resources. Efficiently managing these complexities ensures optimized performance in terms of both pro- cessing time and compression efficiency across varying medical image sizes and ROI dimensions.
The performance of the proposed method was rigorously compared with that of other techniques using similar image configurations. The results, as illustrated in Table 4, unequivocally demonstrate the superiority of the proposed strategy over the alternatives. Furthermore, we conducted a comparative analysis of the Structural Similarity Index (SSIM) values achieved by our method and those obtained through lossless compression techniques employed by other methods. Notably, our approach consistently yielded images with exceptional visual quality, as evidenced by the SSIM values. These findings underscore the effectiveness and superiority of the proposed method in producing high-quality images suitable for visualization purposes.
The effectiveness of the threshold application on the coefficients is assessed after the DWT on the filtered picture and after applying a threshold to the generated coefficients. A two-level decomposition and a three-level decomposition are two separate scenarios that are taken into account. All coefficients below the threshold are set to zero after the transformation. The findings show that there was not too much noise in the image as a result of some coefficients being discarded. Thus, the image's quality is still present. The effect is visible in the high PSNR values. The quantity of coefficients eliminated is quite intriguing. This raises the likelihood of having several chains of zero coefficients. The quantity of discarded coefficients rises with the number of layers.
CONCLUSION
Every image contains some unnecessary information that must be recognized by the user in order to be compressed. Little inaccuracy is introduced into the system by the floating point representation. The DWT's faultless reconstruction property makes it a preferred choice for usage in important medical applications. In addition to better outcomes than lossless techniques, ROI-based compression also preserves data that is crucial for diagnostics. Such a technique is advised for telemedicine systems, particularly in rural areas where network resources are constrained. An enhanced iteration of the proposed method could integrate automatic selection of ROI for compression, based on both information content and image characteristics. In addressing real-time applications for MR DICOM images, consideration of computational complexity is paramount, with the Set Partitioning In Hierarchical Trees (SPIHT) algorithm demonstrating superior performance for ROI-based compression. Additionally, the development of improved recommender systems for large-scale, high-quality medical images will be pursued through the integration of technologies such as the Internet of Medical Things (IoMT), medical imaging techniques, soft computing with evolutionary operators, and various hybrid image processing strategies. These advancements aim to enhance diagnostic capabilities and optimize workflows in medical imaging.
AUTHOR’S CONTRIBUTIONS
It is hereby acknowledged that all authors have accepted responsibility for the manuscript's content and consented to its submission. They have meticulously reviewed all results and unanimously approved the final version of the manuscript.
LIST OF ABBREVIATIONS
| DWT | = Discrete Wavelet Transform |
| RBC | = Region-Based Compression |
| DICOM | = Digital Imaging and Communication in Medicine |
| ROI | = Regions Of Interest |
| MRI | = Magnetic Resonance Imaging |
| CT | = Computed Tomography |
| FFN | = Fully Fused Network |
| ISB | = Improved Single Block |
| DDPM | = Denoising Diffusion Probabilistic Model |
| CBCT | = Cone-beam Computed Tomography |
| 3D-HLCK | = 3-D hierarchical listless clock algorithm |
| 3D-SPECK | = 3-D set partitioning embedded block approach |
| WDR | = Wavelet Difference Reduction |
| MCPD | = Mean Co-located Pixel Difference |
| SSIM | = Structural Similarity index |
| JPEG | = Joint Photographic Experts Group |
| DAC | = Discrete Atomic Compression |
| BP | = Back Propagation |
| MLP | = Multi-Layer Perceptron |
| LSTM | = Long Short-Term Memory |
| SVD | = Singular Value Decomposition |
| CNN | = Convolutional neural networks |
| RBC | = Region-Based Compression |
| RLE | = Run-Length Encoding |
| LZW | = Lempel-Ziv-Welch |
| SPIHT | = Set Partitioning in Hierarchical Trees |
| RGB | = Red, Green, Blue |
| EZW | = Embedded Zerotree Wavelet |
| LIS | = Largest Intensity Sequences |
| LIP | = Insignificant Pixels |
| PSNR | = Peak Signal-to-Noise Rat |
| SNR | = Signal-to-Noise Ratio |
| IoMT | = Internet of Medical Things |


