All published articles of this journal are available on ScienceDirect.
Self-Prompting Hybrid YOLOv12-SAM 2 Model for MRI Brain Tumour Segmentation in Real Time
Abstract
Introduction
The brain is the main organ of the nervous system and serves as the command and control centre for all bodily functions required to maintain a normal, healthy life. Brain tumours are characterised by the growth of abnormal cells in the brain, which disrupts healthy cerebral tissue. Early diagnosis and effective treatment require the timely identification and segmentation of brain tumours. Traditional methods of diagnosing brain tumours include manually reviewing MRIs, which is a laborious and error-prone procedure. Researchers have recently developed many novel automated methods for detecting and segmenting brain tumours in magnetic resonance imaging data. These useful techniques have brought tremendous improvement in the precision and speed of medical image analysis, eventually leading to more accurate diagnoses and optimised treatment plans.
Materials and Methods
In this study, a new method was introduced to use Segment Anything Model 2 (SAM 2) with the YOLOv12 model to detect and segment brain tumour using MRI. In this approach, the predictions of the YOLOv12 bounding box tumour were used to automatically generate input prompts for SAM 2, reducing the need for manual annotations. Then it was applied to a benchmark figshare image dataset where it performed better than the state-of-the-art in the tumor segmentation task.
Discussion
Compared with state-of-the-art models, the proposed model outperforms in terms of segmentation accuracy and the Dice score.
Conclusion
This study indicates that the use of this hybrid model for radiological analysis will significantly increase the accuracy and speed of radiological analysis, with the potential to aid in clinical decision-making and patient care.
1. INTRODUCTION
Brain tumours threaten human life and health, and there is a wide range of treatment options and outcomes based on tumour type. Around 10 million people died from such cases of cancer worldwide in 2020, and there were 19.3 million new cases, highlighting the need for enhanced detection methods [1]. In medical settings, MRI is a commonly used non-invasive diagnostic technique for evaluating brain tumours [2]. Manual brain tumour segmentation takes time, is labour-intensive, and may introduce inter-observer variability, which could prolong diagnosis and treatment planning. Automated brain tumour segmentation is highly reliable, fast, and objective, thus benefiting clinical decision-making and enabling personalised treatments. Recent advancements in computer vision and machine learning have positioned CNNs as a powerful option. These state-of-the-art models have effectively tackled complex computer-aided diagnosis (CAD) challenges, including tasks such as recognition, classification, segmentation, and detection [3]. Despite their success, many current CAD solutions for the detection and segmentation of brain tumours using CNNs are inefficient on various platforms and require significant computational power. This leads to problems when using normal devices, resulting in poor service quality and low performance.
Numerous algorithms have been created for object detection to improve the identification of different objects within images. In 2012, CNNs revolutionised computer vision by learning to extract low- and high-level features from images. Convolutional layers enable CNNs to detect objects irrespective of their position. However, CNNs could not directly handle object detection due to problems, such as the variance in the number of objects, their sizes, and orientations. Object detection requires identifying both class and location using bounding boxes. R-CNN [4], introduced in 2014, used a selective search to generate 2,000 region proposals, processed them through a CNN, and used SVM classifiers for detection. Despite its significance, R-CNN was slow, taking 14 seconds per image with a GPU. Faster R-CNN [5] improved its efficiency by integrating region proposal networks, facilitating end-to-end training, and eliminating the need for external proposal generation. SSD [6] introduced default boxes of various scales, predicted object scores, and adjusted shapes using multiscale feature maps. This approach eliminated separate proposal generation, improving training and small object detection performance.
YOLO (You Only Look Once) improved upon the concepts introduced by R-CNN, Faster R-CNN, and SSD. The original YOLO framework, developed by Redmon et al. [7], has been continuously upgraded. The YOLO algorithm, which consists of a single unified neural network, is highly attractive due to its outstanding object detection capabilities. This algorithm transformed the field of object detection by approaching it as a regression task, directly predicting bounding-box coordinates and class probabilities from pixel-level information. Each new version builds on the previous ones, focusing on enhancing both speed and accuracy. Because of these capabilities, YOLO is used in a wide range of applications, such as medical imaging, high-resolution surveillance systems, healthcare, and manufacturing.
Convolutional neural networks (CNNs) serve as foundational elements in medical image automatic segmentation because of their effective performance in deep learning visual applications. The field of medical image segmentation is largely influenced by U-Net [8], as this approach employs a symmetric encoder-decoder architecture with skip connections to provide dense pixel-level predictions. The U-Net architecture contains two paths: an encoder for dimension reduction and a decoder to restore spatial dimensions, incorporating features from low-resolution layers at each stage. The network implements a U-shaped structure with these two paths. Skip connections within the U-Net allow it to merge contracting path features with expanding path features, enabling the preservation of spatial information.
For better segmentation outcomes, numerous U-Net variants have been further developed, including ResUNet, AttnUNet, and U-Net++, which are based on traditional modules such as dense connections, inception, residual connections, and attention mechanisms. The U-Net architecture is modified by incorporating residual connections in ResUNet, or Residual U-Net [9]. Residual blocks help train deeper networks by addressing the vanishing gradient problem. This also enables the network to learn identity functions, which can improve performance.
Attention U-Net is a variant of the U-Net architecture that incorporates an attention mechanism to improve performance in medical image segmentation [10]. Skip connections are used with attention gates between the encoder and decoder paths. The attention mechanism enables the network to learn which objects it should focus on in an image automatically. Attention U-Net has been applied to various medical imaging tasks, including the segmentation of brain tissues and abdominal structures.
DenseNet served as an inspiration for U-Net++ [11]. This network’s design scheme uses dense blocks and links between the contracting and expanding paths, as well as intermediary grid blocks. In addition to improving segmentation accuracy, these intermediary blocks assist the network in transferring more semantic information between regular paths, especially when their feature maps share strong semantic similarity.
Even with these advances, annotating medical images continues to be a time-consuming and expensive task, as it generally requires medical expertise. This issue has led to a growing interest in transfer learning, which leverages knowledge from extensive natural image datasets for specific medical imaging applications. Recent developments in foundational models, notably the transformer-based model known as the Segment Anything Model (SAM) [12], have demonstrated outstanding performance in creating high-quality object masks from a variety of input prompts. The achievements across multiple computer vision benchmarks have drawn considerable attention for their potential use in medical image segmentation [13].
Previously used AI models in radiology include U-Net, DeepLabv3+, and transformer-based architectures, which have played an important role in enhancing medical image segmentation, but they usually require significant annotated data and lack real-time inference capability. Similarly, detection frameworks such as earlier YOLO variants offer fast localisation but are not optimised for fine-grained segmentation. Although the transformer-based model SAM 2 possesses remarkable zero-shot performance in segmenting medical images [14], it still requires input from human experts. This dependence on manual input limits the efficiency and scalability of the segmentation process.
To overcome this challenge, we propose a self-promoting segmentation model that generates automatic input prompts by utilising the pre-trained capabilities of the YOLOv12 model. Our approach aims to enhance the accuracy and computational efficiency of tumour segmentation by combining YOLOv12 bounding box tumour predictions with the SAM 2 segmentation task. In this work, we use bounding box coordinate data to train the SAM 2 segmentation model. This approach significantly decreases annotation time compared with previous methods that required manual segmentation of ground truth masks for training. By using bounding-box annotations, our model can achieve a high degree of segmentation accuracy with less manual effort, making it more practical for large-scale applications.
The suggested prompt-based brain tumour segmentation model makes the following important contributions.
• In this work, we present a new hybrid model by integrating YOLOv12, a recently powerful object detector, with the SAM 2 framework for specifically performing accurate and fully automated brain tumour segmentation of MRI scans. With this integration, YOLOv12 takes advantage of its real-time tumour localization strength, while SAM 2 takes advantage of its high-resolution segmentation strength, resulting in a complete solution for both detection and segmentation tasks.
• In this research, we present a self-prompt-based mechanism. This work uses bounding boxes produced by YOLOv12 as prompts for segmentation, adequately achieving high accuracy with only bounding-box annotations, thereby reducing annotation time and the expertise needed.
• In this work, the YOLOv12 model is used to detect tumour regions with bounding boxes. Then the bounding boxes are used to guide SAM 2 in segmenting the tumour within the localized area only. This type of targeted segmentation method allocates computational resources to the more important regions, thereby making the model both efficient and accurate.
• The SAM 2 component leverages bounding boxes produced by YOLOv12 to approximate context-aware and precise segmentation through a transformer architecture that generalizes well to different tumour shapes, sizes, and localizations. This collaboration successfully addresses the problem of segmenting tumours with uneven edges, overlapping structures, or low contrast.
• We use the brain tumour dataset from Figshare to assess our suggested model. The hybrid model is also compared with leading segmentation methods based on important metrics, demonstrating that our proposed model is clinically applicable and reliable.
2. EXISTING WORKS
2.1. Related Works
In 2000, Naser et al. [15] designed a CNN-based model using a U-Net architecture for tumour segmentation and detection. The proposed method was capable of tumour segmentation, detection, and classification in a single pipeline using the same MRI data. A tumour classification model was developed using a densely connected neural network classifier, based on a tumour grading model with transfer learning using VGG16 weights. Complete automation becomes possible through this integrated system, which achieves simultaneous processing to improve tumour analysis workflow efficiency.
In 2003, Yousef et al. [16] designed a model based on the lightweight U-Net for brain tumour segmentation. The proposed model uses a much smaller number of trainable parameters (2 million) than the original U-Net (7.7 million). The new model is much more efficient in locating the brain tumour with fewer parameters.
In 2024, Mithun et al. [17] proposed a model based on the YOLONAS deep learning technique for brain tumour classification using a segmentation approach. The proposed hybrid technique involves the combination of an encoder–decoder network with the pre-trained EfficientNet-B3, which acts as the encoder for this model. This architecture aims to improve the segmentation of MRI images by efficiently detecting the features of brain tumours. This approach encompasses several stages, namely image pre-processing, segmentation, and classification.
In 2022, Ottom et al. [18] presented a new Znet framework for the segmentation of 2D brain tumours. It employs deep neural networks as well as data augmentation strategies. Znet has been designed based on skip connections and an encoder–decoder architecture. The Znet model has exhibited excellent results with a Dice coefficient on an independent testing dataset.
In 2025, Ahsan et al. [19] designed a model combining YOLOv5 with 2D U-Net for multiclass tumour analysis. In this model, YOLOv5 was used for detecting tumour localization, and U-Net was used for segmenting the tumour. This model decreases the amount of time needed for diagnosis and thus facilitates early treatment.
In 2022, Sami et al. [20] proposed a modified U-Net method. Traditional U-Net methods have errors in accurately identifying tumours. The modified U-Net architecture differs from the traditional U-Net architecture. Compared to the original U-Net, the modified U-Net has six convolutional layers, replaces 3×3 filters with 5×5 filters, uses 2,048 feature maps, and reduces the feature maps to 8×8 in the encoding part. These modifications allow the model to increase feature map dimensionality, resulting in improved performance.
In 2024, Saifullah et al. [21] presented a CNN with a transfer learning model. In this model, DeepLabv3+ with a ResNet18 backbone was used for tumour segmentation. DeepLabv3+ is highly effective in semantic segmentation, which is essential for accurately defining object boundaries in medical images. ResNet18 serves as the feature extractor (backbone) within DeepLabv3+. This strategic fusion improves performance in the challenging task of tumour segmentation.
In 2023, Nizamani et al. [22] introduced a deep learning model that combined the U-Net architecture with a transformer model and also used advanced feature enhancement techniques. Transformers were added to the U-Net model to impart contextual understanding, allowing the model to better capture the relationships between different parts of the image, which is crucial for accurate segmentation. Feature enhancement techniques in image pre-processing, such as MHE, CLAHE, and MBOBHE, were used to improve the visibility of important details. These techniques improved tumour segmentation accuracy.
In 2024, Zafar et al. [23] designed a hybrid deep learning model called Enhanced TumorNet. In this model, YOLOv8s and U-Net are combined for improved tumour analysis. YOLOv8s is used for rapid detection, and U-Net is used for precise segmentation. The overall performance of this model is improved by utilising the advantages of both architectures.
In 2024, Kassam et al. [24] proposed a model that combines YOLOv8 and a Segment Anything Model (SAM) to improve the analysis of glioma tumours. The YOLOv8 model provides quick and efficient processing of MRI images through a CNN to detect and localise potential tumours by predicting bounding boxes, while SAM offers precise segmentation, creating a robust and efficient pipeline.
Table 1 presents the features and challenges of traditional and existing brain detection and segmentation models.
| Author | Methodology | Features | Challenges |
|---|---|---|---|
| Naser et al. [15] | U-Net+Vgg-16 | 1. The data imbalance was addressed by the authors using a weighted loss function. 2. The method offers a noninvasive way to characterize lower-grade cancers, which may help with diagnosis and therapy planning. |
1. In this study, manually segmented tumour masks were used as the basis for training the U-Net model. 2. Although the U-Net is good at capturing local features, it may struggle to capture long-range dependencies, which could be important for accurately segmenting complex tumour structures. |
| Yousef et al. [16] | lightweight U-Net | 1. Significantly reduces computational complexity. 2. The use of the cyclical learning rate(CLR) leads to a better generalisation in segmenting brain tumors. |
1. The model may not perform equally well in different data distributions or tumour types. |
| Mithun et al. [17] | Yolo NAS | 1. YOLO NAS prioritises important areas in the images using an attention method. 2. The pre-processing step includes noise reduction using a hybrid anisotropic diffusion filtering technique, which enhances the quality of MRI images before analysis. |
1. Some segmented images may contain small fragments incorrectly identified as tumours. 2. The segmentation results depended on the HADF technique. |
| Ottom et al. [18] | Znet | 1. Potentially avoids the vanishing gradient problem. 2. Potential of the architecture for clinical applications. |
1. The Znet model has more trainable parameters (44,384,833) than the U-Net model (14,788,929), which requires higher computation. 2. The performance of Znet in other MRI sequences or combined modalities has not yet been explored. |
| Ahsan et al. [19] | Yolov5+U-Net | 1. YOLOv5 and 2D U-Net together produced a higher DSC for segmentation than 2D U-Net alone. 2. Less inference time is required to detect tumors. |
1. The proposed method requires the training of two separate models, which can be more computationally intensive than single model approaches. |
| Sami et al. [20] | Modified U-Net | 1. The modified U-Net includes additional layers and changes in filter size, which contribute to its superior performance. | 1. The modified U-Net has more layers and larger filter sizes, which likely increases computational requirements. 2. A manual annotation mask was required to train the model. |
| Saifullah et al.[21] | Deeplabv3+ + ResNet18 | 1. Atrous Spatial Pyramid Pooling enhances the ability of the model to recognise both large and small tumor regions. 2. The excellent performance of the model in defining tumour boundaries. |
1. Computationally intensive models. 2. Its applicability in resource-constrained clinical settings. |
| Nizamani et al. [22] | U-Net+Transformer | 1. Transformer-based models can effectively extract contextual and spatial information from magnetic resonance data, which helps in segmentation of tumors with irregular shapes. |
1. The integration of U-Nets with Transformers and advanced feature enhancement techniques likely results in a computationally intensive model. |
| Zafar et al. [23] | Yolov8’s+U-Net | 1. The model is suitable for real-time applications. | 1. The hybrid model that combines YOLOv8s and U-Net is computationally intensive. 2. This model requires high-quality data to achieve optimal performance. |
| Kassam et al. [24] | Yolov8+SAM | 1. This pipeline is suitable for real-time application. 2. The model provides a low inference time to detect and segmenting tumors. |
1. This supports only two-dimensional (2D) MRI tumour segmentation. |
2.2. Research Gap
The research gaps mentioned below were found in the literature review.
· For training, traditional segmentation algorithms, such as U-Net and DeepLabv3+ require ground-truth masks; however, the process of constructing these masks is challenging because healthcare professionals must manually annotate them. This process is time-consuming and requires specialised radiological knowledge, restricting scalability.
· Conventional models, such as U-Net and DeepLabv3+, attempt to segment the entire MRI image at once without first identifying the tumour, which can reduce precision in complex-structure tumours or noisy images.
· Slow inference times are a common problem with conventional high-accuracy segmentation algorithms, making them inappropriate for real-time or mobile health applications.
· There is still much to explore regarding the potential of combining prompt-based segmentation techniques, such as the transformer-based SAM2, with high-performance detection models, such as YOLOv12, in brain tumour applications.
3. PROPOSED METHODOLOGY
Our proposed model combines two cutting-edge algorithms, YOLOv12 and SAM 2, to efficiently identify and segment brain tumours in MRI images. Initially, the YOLOv12 model detects the location of tumours and encircles them with bounding boxes. Subsequently, these bounding boxes are used as input prompts for the SAM 2 model, which accurately segments the tumour using the given bounding box coordinates (Fig. 1). A pre-trained YOLOv12 model is used for tumour detection. This model is selected for its remarkable accuracy and speed performance, especially in real-time applications.
YOLOv12 uses a CNN to process MRI images. This CNN extracts essential features from the images, and based on these features, YOLOv12 predicts bounding boxes around potential tumours. The coordinates of the bounding boxes predicted by YOLOv12 are then used as input for the SAM 2 model. To handle the output of YOLOv12 in our environment, a conversion mechanism is created that translates the coordinates of the bounding boxes into spatial prompts that SAM 2 can handle properly. Bounding boxes are rescaled to fit the input dimensions of SAM 2, and other preprocessing operations are applied to handle tumours with irregular or overlapping boundaries. This smooth transition between the detection and segmentation processes reduces the dependence on manual annotations and increases both precision and efficiency. These coordinates serve as prompts for SAM 2, guiding its segmentation process.
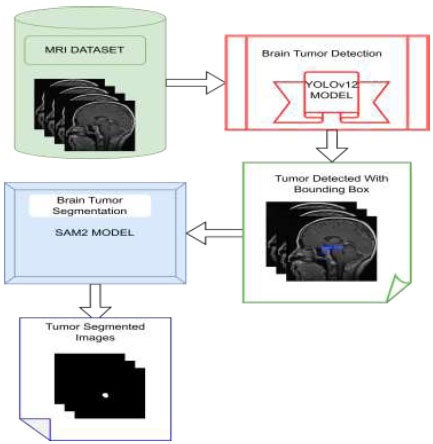
The suggested model's workflow.
The SAM 2 model, lightweight and highly accurate, refines YOLOv12's detection results. It uses bounding-box coordinates to perform detailed segmentation, delineating exact tumour boundaries.
3.1. Dataset
The dataset used in this study was the publicly available Brain Tumour MRI Figshare dataset, originally hosted on FigShare and also available on Kaggle for accessibility [25]. It contains 3,064 T1-weighted contrast-enhanced MRI images and their corresponding binary masks. The image size of the MRI dataset was 640×640. The dataset was divided into 2,451 training images, 307 validation images, and 306 test images. For visual inspection, a random selection of MRI images was chosen, as shown in Fig. (2A), to guarantee the consistency and high quality of our data. Fig. (2B) shows these images superimposed with the corresponding ground-truth masks. This step is crucial to verify that the MRI images and ground-truth masks are properly aligned for training our prediction model. Annotation text labels are required to train YOLOv12; therefore, the binary masks were converted to label.txt files for training YOLOv12.
3.2. Data Augmentation
The data augmentation techniques used during the training of the YOLOv12 model included the following:
• Blur: Applying a slight Gaussian blur with a probability of 0.01. This enhances the model's ability to handle motion blur or low-quality images during inference.
• MedianBlur: Using median blurring, also with a probability of 0.01. This alteration helps the model manage noise or sharp edges present in the images.
• ToGray: Converting images to greyscale, with a probability of 0.01, enabling the model to concentrate on spatial characteristics instead of colour, which can be advantageous when colour information is less significant or unreliable.
• CLAHE: Improving image contrast with a probability of 0.01, assisting the model in focusing on key features by enhancing the visual distinctiveness of the images.
These enhancements were chosen to simulate real-world variations in object detection tasks, such as changes in lighting, image quality, and environmental factors that can affect object visibility and appearance. The augmentation objective was to decrease overfitting and improve the models' capacity for generalization.
3.3. Demonstration of the Proposed YOLOv12 for Detecting Brain Tumours
YOLOv12 demonstrates remarkable improvement in the field of real-time object detection by expanding the main concepts of the YOLO model series. YOLOv12 was launched in February 2025 by researchers Tian et al. [26]. In the YOLOv12 model, attention mechanisms are added to improve detection precision without affecting its rapid processing capabilities. In the proposed model, YOLOv12 is used for tumour detection with bounding boxes. Several important changes have been introduced in the architecture of YOLOv12 to detect objects more accurately. The architecture of YOLOv12, presented in Fig. (3), contains three main components [27].
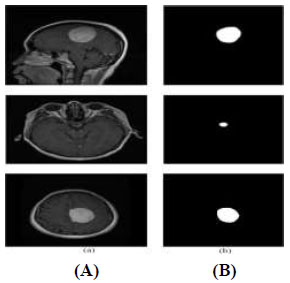
Sample images of the dataset: (A) MRI images and (B) corresponding ground-truth masks.
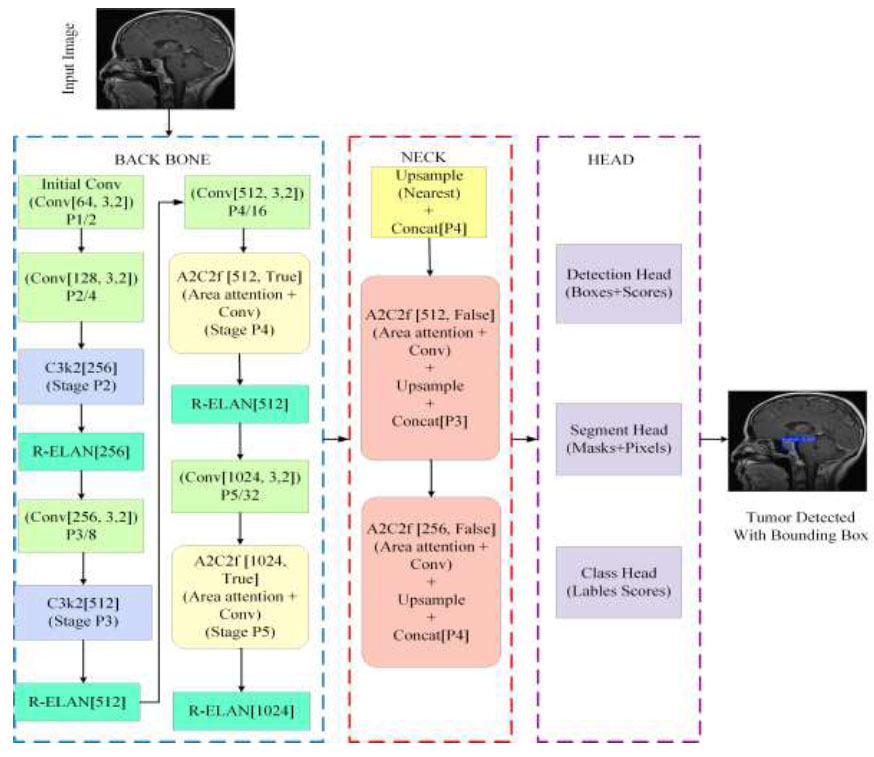
Architecture of YOLOv12.
3.3.1. Backbone
YOLOv12 uses a Residual Efficient Layer Aggregation Network (R-ELAN) as its backbone. This network is designed with deeper residual connections to improve feature extraction and reuse. It uses 7×7 separable convolution layers to capture spatial context while limiting the number of trainable parameters. Advanced convolutional blocks, which are key components of the R-ELAN, are employed in the backbone to enhance feature extraction while maintaining computational efficiency [28]. These strategies improve performance without increasing computational overhead. By using these lightweight operations with higher parallelization in the backbone, YOLOv12 achieves faster processing speeds for real-time object detection.
A novel convolutional block introduced by YOLOv12 aims to reduce the complexity of the operation and increase parallelism, allowing it to perform better than its previous versions. These blocks are divided into a sequence of smaller kernels, which can be represented by Eq. (1).
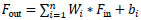
Where Fout is the output featuremap, Wi is the convolutional filter,Fin is the input feature
Instead of using a few larger convolutions, YOLOv12 processes information more quickly by distributing the computing among many compact convolutions.
3.3.2. Neck
YOLOv12 connects its backbone to the head through the neck. It also incorporates multi-scale features from various regions of the network. An area-attention mechanism helps the model focus on challenging details in the image. The area attention builds on the FlashAttention network, which significantly reduces memory and computational costs. It aggregates information from multiple scales, refines it, and then passes it to the head for predictions. The process of the attention operation in the neck is described mathematically in Eq. (2).
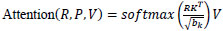
Where R, P, and V are query, key, and value matrices, and is the dimensionality of the key.
3.3.3. Head
In YOLOv12, the head receives information from the neck and generates the final outputs, such as the bounding box coordinates and the class of each box. It uses loss functions that balance localization and classification objectives, resulting in improved overall detection performance. Both the loss functions and the prediction pathways are designed for fast and efficient operation in real-time applications. YOLOv12 may employ an extended version of the typical YOLO-style loss function, which is mathematically represented by Eq. (3).
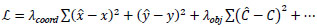
Where
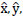 and
and
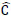 denote predicted,bounding box coordinates,and confidence,respectively.
denote predicted,bounding box coordinates,and confidence,respectively.
These architectural components work together to achieve a balance between computational efficiency and detection accuracy. YOLOv12 also introduces multiple model versions (e.g., 12n, 12s, 12m, 12x), allowing users to prioritise speed or accuracy based on their specific requirements.
3.4. Demonstration of the proposed SAM2 model for Detecting Brain Tumour Segmentation
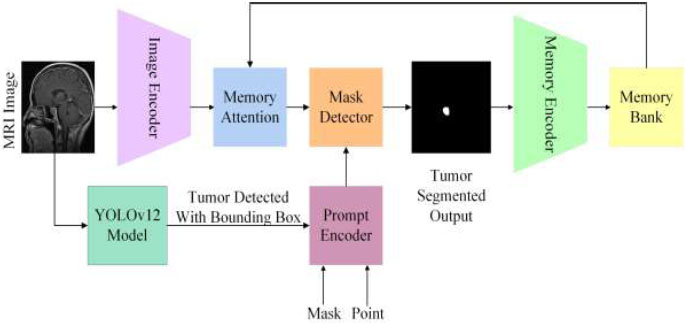
Segment Anything Model (SAM) is a foundation model designed for image segmentation that can be initiated through prompts [29]. SAM 2 was created for prompt-based visual segmentation in both images and videos, expanding SAM's capabilities to include the video domain. SAM 2 demonstrates better performance in medical image segmentation while being significantly faster than the original SAM [30]. The proposed hybrid model employs SAM 2 for segmenting brain tumours, using the input prompt derived from YOLOv12's brain tumour detection with a bounding box, as shown in Fig. (4). SAM 2 contains the following three components for image segmentation [31].
3.4.1. Image Encoder
SAM 2 uses a Hiera image encoder pre-trained with MAE (Masked Autoencoder). For single images, this encoder processes the input and provides unconditioned tokens (feature embeddings) that represent the spatial and semantic details of the image.
SAM 2 Architecture
3.4.2. Prompt Encoder
The prompt encoder is designed to handle different user inputs, such as points, boxes, or text, to guide the segmentation process. It interprets these prompts and transforms them into a feature space corresponding to the image features derived by the image encoder.
3.4.3. Mask Decoder
The mask decoder uses two-way transformer blocks to update prompt and image embeddings. Its role is to combine the features from the prompt encoder and the image encoder to produce the optimal segmentation prediction. The two-way transformer blends these features and incorporates an IoU head to assess the quality of the segmentation mask.
3.5. Model Training, Fine-Tuning, and Transfer Learning
The SAM 2 models were used in their pre-trained zero-shot large (ViT-L) versions, utilising default settings for inference without any fine-tuning or training. The input prompts for SAM 2 were generated by YOLOv12. Thus, only the YOLOv12 model was trained to identify brain tumours, while the SAM 2 model remained frozen and was used solely for segmentation tasks. This strategy leverages YOLOv12's detection process and relies on the pre-trained segmentation capabilities of SAM 2 without updating its original weights. The fine-tuning of the hyperparameters of the YOLOv12 model is presented in Table 2.
4. RESULTS AND DISCUSSION
4.1. Environmental Setup
The experiments were conducted using the Google Colab Pro+ platform, which provides a GPU-accelerated environment. The hardware configuration included an NVIDIA Tesla T4 GPU, offering 15,360 MiB of dedicated memory. The system was configured with NVIDIA drivers version 550.55.17 and CUDA version 13.4. The back-end environment of Google Colab was based on Ubuntu 20.04 and utilised Python 3.10.12.
4.2. Metrics
To evaluate the quality of the suggested model segmentation, we employed pixel-level metrics including accuracy, Dice coefficient, IoU, precision, recall, and F1 score. Table 3 represents the definitions of the metrics.
| S.No. | Parameter | Setting |
|---|---|---|
| 1. | Model Architecture | YOLOv12m (497 layers, 2,519,859 parameters, 6.0 GFLOPs) |
| 2. | Transferred Weights | 499/499 items transferred from pre-trained weights |
| 3. | Dataset | Figshare Brain Tumour MRI dataset (3,064 images) |
| 4. | Data Split | 80:10:10 |
| 5. | Pre-processing: Data Augmentation | Blur, MedianBlur, ToGray, CLAHE (probabilities = 0.01 each), CopyPaste (0.1), RandAugment |
| 6. | Optimizer | AdamW (learning rate = 0.002, momentum = 0.9) |
| 7. | Batch Size | 16 |
| 8. | Input Image Size | 640x640 pixels |
| 9. | Epochs | 75 |
| 10. | Optimizer Configuration | Weight decay: 0.0005 (128 groups), 0.0 (121 groups), bias: 0.0 (127 groups) |
| 11. | Loss Functions | CIoU loss (bounding box regression), focal loss (classification), and Cross-entropy (segmentation refinement) |
| 12. | Bounding Box to SAM2 Prompt Conversion | YOLOv12 bounding boxes (xmin,ymin,xmax,ymax) normalised to SAM2 input resolution. |
| 13 | Baselines Compared | U-Net, DeepLabv3+, YOLOv8+SAM |
| S.No. | Metrics | Definition |
|---|---|---|
| 1. | Dice Score |
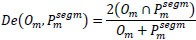 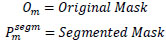
|
| 2. | Accuracy |
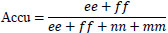 [32] [32]ee: True Positives, ff: True Negatives, nn: False Positives, mm: False Negatives |
| 3. | Precision |
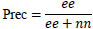
|
| 4. | Recall |
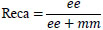
|
| 5. | F1-Score |
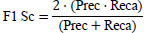 [33] [33]
|
| 6. | IOU |
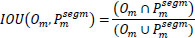 |
4.3. Pseudocode for Tumour Segmentation
Pseudocode for tumour Segmentation is presented in Table 4. Combining YOLOv12 detection and SAM 2’s segmentation allows the proposed framework to achieve accurate tumour segmentation. Initially, the YOLOv12_ TUMOR_DETECTION function processes the medical image using the YOLOv12 model and pinpoints potential tumour regions, along with information about their class identifiers, confidence score, and the coordinates of the bounding box. Each detected region of interest (ROI) is then passed to the SAM 2 segmentation pipeline. For each bounding box, the SAM2_SEGMENTATION function is run to produce a detailed segmentation mask by setting binary values over the region of the bounding box.
| S.No. | Code |
|---|---|
| 1. | Function HYBRID _TUMOR_SEGM(image, yolov12_model, sam2_model) |
| 2. | Detections YOLOv12_TUMOR_DETECTION(image, yolov12_model) |
| 3. | Tumor_info ← [ ] |
| 4. | For Each detection in the detections: do |
|
5. |
Class_id detection[“class_id-tumour”] |
|
6. |
Confidence detection [“confidence Score"] |
|
7. |
Bounding_box ← detection["bounding_box coordinates"] |
|
8. |
Predictor INIT_SAM2(sam2_model) |
|
9. |
Predictor.set_image(image) |
|
10. |
Segmentation_mask SAM2_SEGMENTATION(predictor, bounding_box) |
|
11. |
Tumor_infom.append({ |
|
12. |
"Class_id": class_id-tumour, |
|
13. |
"Confidence": detected confidence score, |
|
14. |
"Bounding_box": detected bounding_box, |
|
15. |
"Segmentation_mask": segmentation_mask |
|
16. |
}) |
|
17. |
End for |
|
19. |
Return tumor_infom |
|
20. |
End function |
|
21. |
Function YOLOv12_TUMOR_DETECTION(image, model) |
|
22. |
Results model(image) |
|
23. |
Extract detections with class_id, confidence, bounding_box |
|
24. |
Return a list of detections |
|
25. |
End function |
|
26. |
Function INIT_SAM2(model) |
|
27. |
Predictor SamPredictor(model) |
|
28. |
Return predictor |
|
29. |
End function |
|
30. |
Function SAM2_SEGMENTATION(predictor, bounding_box) |
|
31. |
x_min, y_min, x_max, y_max bounding_box |
|
32. |
Box [x_min, y_min, x_max, y_max] |
|
33. |
Masks, _, _ predictor.predict(box=box[None, :]) |
|
34. |
Segmentation_mask ← (masks[0] > 0.5) |
|
35. |
Return segmentation_mask |
|
36. |
End function |
4.4. YOLOv12 Model Outcomes of Tumour Detection
Various performance metrics of tumor detection using YOLOv12 model are shown in Fig. (5), highlighting several aspects of the model during training and validation phases over 75 epochs. During the training phase, both box loss and classification loss decreased as the number of epochs increased, indicating progressive improvement in the model’s classification capabilities. The dfl_loss is employed to increase the accuracy of bounding-box predictions, particularly for objects that are difficult to differentiate or closely resemble each other. During the model training, df_loss decreased as the number of epochs increased, demonstrating that the model accurately predicted the bounding boxes. Durning the validation, box_loss, cls_loss, and dfl_loss decreased sharply as the number of epochs increased. Finally, our YOLOv12 model achieved a precision of 90.28%, a recall of 86.66%, and an mAP50 of 92.9%. Thus, the resulting model nearly perfectly predicted tumour detection.
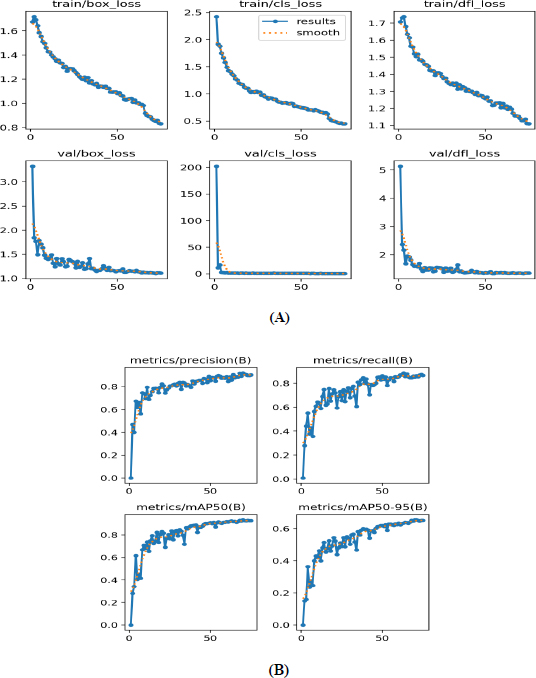
Performance metrics of the YOLOv12 model. (A) Loss curves; (B) Precision, recall, and mAP50 curves.
The confusion matrices associated with the suggested YOLOv12 model are graphically depicted in Fig. (6) using the validation data. The normalised confusion matrix is utilized to pinpoint the highest and lowest performing classes, providing a comprehensive understanding of the model's performance across various categories.

YOLOv12 model normalised confusion matrix.
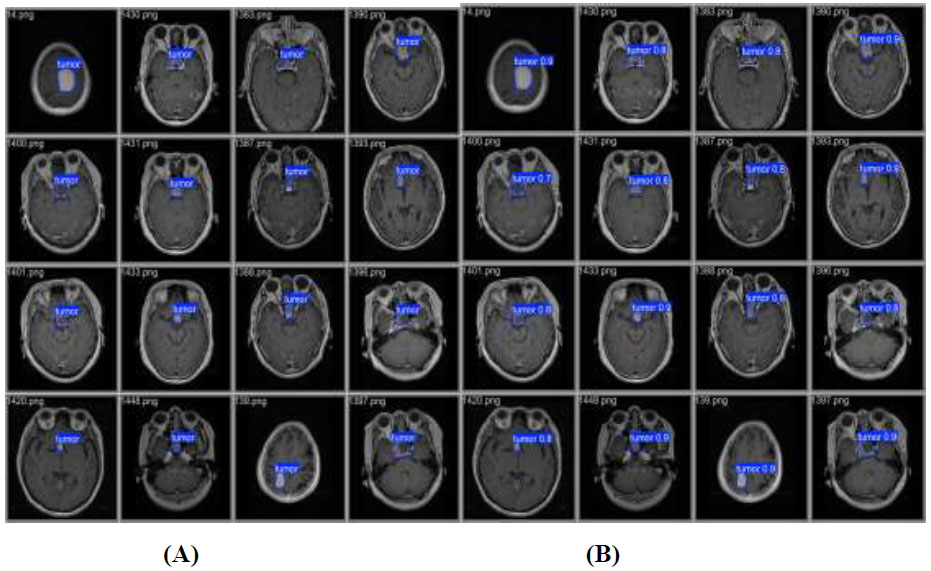
Validation and prediction batches for tumour detection (A) validation batch and (B) predication batch.
presents a visual comparison between the validation batch (Fig. 7A) and the prediction batch (Fig. 7B) generated by the YOLOv12 model. These batches are designed to improve computational efficiency and increase inference speed, facilitating the detection of tumors across various images. The validation images display tumor locations on the original images, whereas the predicted images show the model’s output after training. From this comparison, it can be concluded that YOLOv12 exhibits exceptional tumor detection performance, as evidenced by the accurate localization of tumors with bounding boxes and the corresponding confidence scores, closely aligned with the validation batch.
4.5. Results of the Hybrid Proposed Model
The YOLOv12–SAM 2 framework was evaluated by comparing the predicted segmentation masks with the corresponding ground truth masks. In Fig. (8), several test cases are presented to illustrate the model’s performance in segmenting brain tumours. The figure shows that the predicted masks closely match the ground truth masks, indicating that our system can accurately and effectively segment brain tumors.
Segmentation quality was evaluated using key performance metrics, including Dice Coefficient (DSC), Intersection over Union (IoU), accuracy, recall, precision, and F1 score. The segmentation results are illustrated in Fig. (9) using these metrics. Our model achieved an accuracy of 99.73%, a recall of 89.06%, a precision of 94.75%, a DSC of 91.8%, an F1 score of 91.82%, and an IoU of 84.87%. These results demonstrate that the proposed model provides highly accurate and reliable segmentation of brain tumours.
The results, obtained using a fixed split of 2,451 training images, 307 validation images, and 306 test images, show consistent performance across multiple evaluation metrics. In addition to accuracy and Dice coefficient, other measures such as precision, recall, F1 score, and IoU were also calculated and are presented with their mean, median, and standard deviation to reflect both central tendency and variability, as shown in Fig. (10). This comprehensive assessment confirms the robustness of the developed model and provides further confidence in its generalizability.

Some test cases of ground truth mask vs. predicted mask.
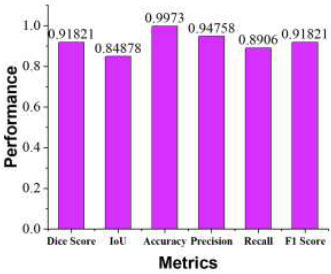
Proposed model metrics vs. performance.
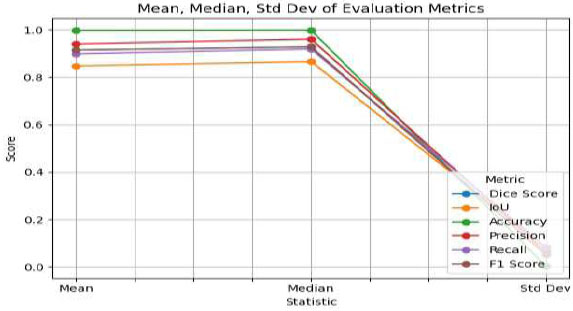
Mean, median, and standard deviation metrics of proposed model.
4.6. Heatmap Overlay
The segmented output mask is a plain black-and-white image and is separate from the original MRI scan, making it difficult to visualize the actual tumour area on the MRI. A heatmap overlay, which adds color to the tumour region, can be blended on top of the original MRI image, as shown in Fig. (11). This heatmap overlay helps non-technical AI users, such as doctors, quickly identify the predicted tumour region within the original MRI scans.
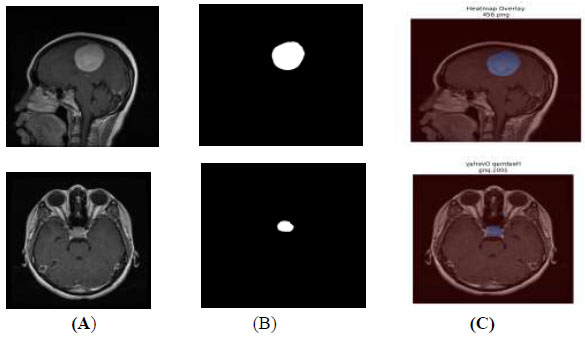
Heatmap overlay (A) original image (B) segmented output (C) heatmap overlay.
4.7. Comparison of the Proposed Model with Existing Models
In this section, we conduct a comprehensive evaluation of the performance of our integrated YOLOv12-SAM 2 model in tumour segmentation. The model’s remarkable accuracy and capability in segmenting tumours are underscored by performance metrics, including a Dice coefficient of 91.8% and an accuracy of 99.7%. Our model outperforms alternative techniques, such as U-Net, DeepLabv3+, and other YOLO models, as indicated in Table 5. The Dice coefficient of 91.8% reflects a precise spatial overlap between the predicted masks and the ground-truth masks. Additionally, the 99.7% accuracy score highlights the model’s reliable ability to detect and segment tumours. YOLOv12 provides accurate localisation through bounding boxes, which are then used as prompts for SAM 2 to enable segmentation in regions with complex tumour boundaries. This combination of complementary detection (YOLOv12) and segmentation (SAM 2) mechanisms leads to more robust results, with fewer false positives and better generalisation across a wide range of MRI scans.
Although alternative methods, such as YOLONAS, YOLOv8 + U-Net, DeepLabv3+ + ResNet18, 2D-UNet, and Znet demonstrate commendable performance metrics, our proposed approach consistently surpasses them in terms of the Dice coefficient, highlighting its superior accuracy in tumour segmentation. The effectiveness of our method is particularly evident when compared to Znet, producing comparable outcomes in both accuracy and Dice coefficient. The high accuracy and Dice score demonstrate the model’s precision in segmenting and delineating tumour regions. These findings indicate that our model is a powerful tool in neuroradiology, capable of diagnosing brain tumours with greater precision, especially in complex cases that are challenging for human evaluation.
Figure (12) represents the comparative performance of YOLOv12+SAM2 and existing state-of-the-art models in terms of precision and Dice coefficient.
| S.No. | Model | Accuracy | DICE Score |
|---|---|---|---|
| 1. | 2D-Unet [34] | 92.16 | 81.2 |
| 2. | MSD [35] | - | 84.69 |
| 3. | 3D-Unet [36] | - | 86 |
| 4. | YOLONAS [17] | 96.20 | 85.81 |
| 5. | Znet [18] | 99.55 | 91.58 |
| 6. | YOLOv5 + 2D U-Net [19] | - | 88.1 |
| 7. | Modified U-Net [20] | 99.5 | 85.02 |
| 8. | Deeplabv3+ +ResNet18 [21] | 97.48 | 91.2 |
| 9. | Yolov8+U-Net [23] | 98.6 | - |
| 10. | Yolov8+SAM [24] | - | 79 |
| 11. | Proposed Model | 99.7 | 91.8 |
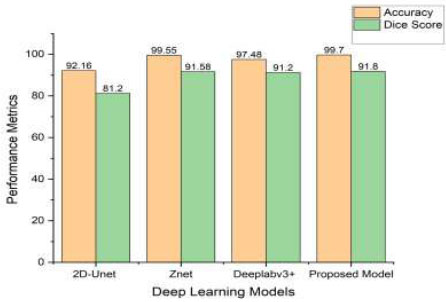
Comparative performance of YOLOv12+SAM2 and existing state-of-the-art models.
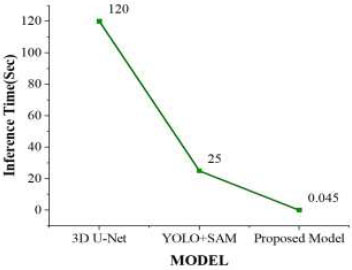
Inference times of the tumour segmentation model.
4.8. Detection Time
The evaluation of the models was carried out on a dataset comprising 91 frames, each with an image dimension of 640 × 640 pixels. Figure (13) represents the inference time for different models. The inference time for the 3D U-Net model was approximately 2 minutes to complete the tumour segmentation process, while the YOLO + SAM model required 15 to 25 seconds. In contrast, our proposed model required only 40–45 milliseconds to segment the tumour. Table 6 shows a comparison of inference times for different state-of-the-art models. Due to its significantly reduced inference time, the YOLOv12 + SAM 2 model was better suited for practical MRI brain tumour segmentation applications, providing faster and reliable outcomes during real-time surgical procedures.
| S.No. | Model | Inference Time(sec) |
|---|---|---|
| 1. | 3D U-Net [37] | 120 |
| 2. | YOLO+SAM [24] | 25 |
| 3. | Proposed Model | 0.045 |
5. LIMITATIONS AND FUTURE WORK
This model performed better on large or regularly shaped tumours, but the dataset used may introduce biases due to limited diversity in patient populations and MRI acquisition conditions. Furthermore, the current evaluation is restricted to conventional MRI sequences, which may limit the generalisability of the results to other imaging modalities and tumour subtypes. The clinical applicability and reliability of the proposed model in practical diagnostic settings could be further enhanced by diversifying and expanding the dataset to include multimodal and heterogeneous magnetic resonance images. Additionally, incorporating radiologist feedback loops would allow continuous refinement of the model to better align with expert clinical practice.
CONCLUSION
In this paper, we presented a self-prompting brain tumour segmentation model that combines the advantages of SAM 2 and YOLOv12 for real-time brain tumour detection and segmentation. Our method overcomes the drawbacks of manual input prompts by using YOLOv12’s pre-trained features to produce bounding box predictions, which SAM 2 then uses for precise segmentation. Through extensive experiments with brain tumour datasets, we showed that our model outperforms current state-of-the-art techniques. The notable enhancements in segmentation accuracy, along with the decreased requirement for detailed ground-truth masks, underscore the practicality and efficiency of our approach for large-scale applications. The model achieved a DICE score of 91.8%, an accuracy of 99.7%, and an inference time of 45 milliseconds, demonstrating strong capability for efficient and effective tumour segmentation. This advancement could significantly impact the field of tumour surgery, as incorporating this model with an intraoperative MRI (ioMRI) system may lead to enhanced patient outcomes and more successful surgical procedures. To achieve clinical translation, the data should be revalidated with radiologists to further confirm the validity of the segmented outputs. Furthermore, practical issues, such as hardware limitations and inference lag, must be addressed prior to integration into radiology workflows. Future steps toward deployment will thus require an iterative approach of testing with clinical specialists to reinforce real-world applicability.
AUTHORS’ CONTRIBUTIONS
The authors confirm their contributions to the paper as follows: K.C.P.: was responsible for study conception and design; C.h.R.P.: was responsible for the analysis and interpretation of results; K.R.K.: was responsible for drafting the manuscript; and all authors reviewed the results and approved the final version of the manuscript.
LIST OF ABBREVIATIONS
| DL | = Deep Learning |
| CAD | = Computer Aided Diagnosis |
| SAM2 | = Segment Anything Model2 |
| YOLO | = You Only Look Once |
| MRI | = Magnetic Resonance Imaging |
| SSD | = Single Shot Multibox Detector |
| CNN | = Convolutional Neural Networks |
| RCNN | = Region-Based CNN |
AVAILABILITY OF DATA AND MATERIALS
The data supporting the findings of the article is available in the Brain tumor dataset at [https://figshare.com/articles/ dataset/brain_tumor_dataset/151242 7] [25].
ACKNOWLEDGEMENTS
The authors would like to acknowledge SR University, Warangal, for providing the necessary infrastructure for this research.


