All published articles of this journal are available on ScienceDirect.
Oral COVID-19 Disclosing Test: A Novel Rapid Technique in Infection Diagnosis
Abstract
COVID-19 is an acute respiratory disease caused by novel coronavirus SARS-CoV-2 or 2019-nCoV. Recently, on March 11, 2020, COVID-19 was declared by the WHO as a virus pandemic disease. Nucleic acid real time polymerase chain reaction (PCR) test has become the standard method for diagnosis of SARS-CoV 2 infection; these real time PCR test kits have many limitations. Antibody tests are expensive and not available, especially in the developing countries. There is an urgent need for an accurate and rapid test method to quickly identify a large number of infected patients and asymptomatic persons, and also which can be available all over the world. We propose a new test technique based on the use of oral gel, mouthwash, or tablets that color the area where the virus is localized in mouth, to diagnose the COVID-19 infection. In fact, our test is composed of specific COVID-19 antibody IgM and IgG coupled to colorful or fluorescent molecules. As of April 2020, the study is waiting to be funded and clinical trials will be prepared to be lunched to get advantage of the technique in order to improve COVID-19 testing.
Viral diseases continue to emerge and represent a serious issue to public health. In the last twenty years, several viral epidemics have been recorded such as the severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002 to 2003, and H1N1 influenza in 2009. The last epidemic caused by the Middle East respiratory syndrome coronavirus (MERS-CoV) was first identified in Saudi Arabia in 2012 [1, 2].
The 2019-nCoV, also called SARS-CoV-2, was first reported in Wuhan, China in December 2019. The disease was later named Coronavirus Disease 2019 (COVID-19) and the virus responsible for it as the COVID-19 virus, respectively, by the World Health Organization (WHO) [3, 4].
COVID-19 is an acute respiratory disease caused by novel coronavirus SARS-CoV-2 or 2019-nCoV. Recently, on March 11, 2020, COVID-19 was declared by the WHO as a virus pandemic disease [5, 6].
CoVs are positive-stranded RNA viruses with a crown-like appearance under an electron microscope (coronam is the Latin term for crown) due to the presence of spike glycoproteins on the envelope. Genomic characterization has shown that probably bats and rodents are the gene sources of alphaCoVs and betaCoVs. On the contrary, avian species seem to represent the gene sources of deltaCoVs and gammaCoVs [1, 7, 8].
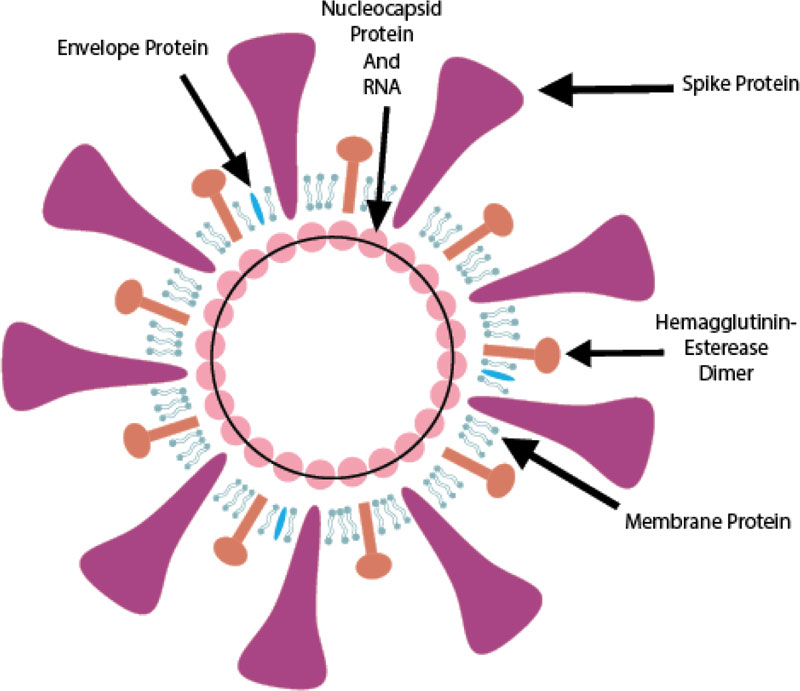
SARS-CoV-2 is from the betaCoVs viruses and has round, elliptic, or pleomorphic form with a diameter of almost 60-140 nm (Fig. 1).
It is sensitive to ultraviolet rays and heat, and maybe inactivated by lipid solvents as ether, ethanol, chlorine-containing disinfectant, peroxyacetic acid and chloroform [1].
(1) The glycosylated Spike protein (S) is a large component making the distinct spikes on the surface of the virus. It utilizes an N-terminal signal sequence to gain access to the endoplasmic reticulum and mediates the attachment to host angiotensin-converting enzyme II (ACE2) receptors. The S protein is cleaved by a host cell furin-like protease into two separate polypeptides S1 and S2.
(2) The RNA genome of the virus is bound by the phosphorylated nucleocapsid protein (N) in a beads-on-a-string conformation. On entering the host cells, the N protein potentially tethers the viral genome to replicase-transcriptase complex (RTC), and helps in packaging the encapsulated genome into viral particles.
(3) The Envelope protein (E) is found in small quantities in the virus and appears to be a transmembrane protein with ion channel activity. The protein facilitates the assembly and release of new virions. It is related to disease pathogenesis and is important for disease progression.
(4) The Membrane protein (M) is the most abundant structural component of the virus. It exists as a dimer and maintains the membrane curvature on one end and binds to nucleocapsid proteins on the other.
(5) The Hemagglutinin-esterase (HE) is also a dimer protein and binds to sialic acids on surface glycoproteins. It is responsible for facilitating and enhancing S protein-mediated cell entry and virus spread through the mucosa [9].
As with other respiratory pathogens, including flu and rhinovirus, the transmission is believed to occur through respiratory droplets from coughing and sneezing or by being carried to the oral or nasal mucosa by hands from the virus-infested surfaces. It appears that all COVID-19 patients – asymptomatic, mild or severe have a massive throat/mucus titre of the virus, shedding it in the surroundings. Aerosol transmission is also possible in case of protracted exposure to elevated aerosol concentrations in closed spaces. Analysis of data related to the spread of SARS-CoV-2 in China seems to indicate that close contact between individuals is necessary. The spread, in fact, is primarily limited to family members, healthcare professionals, and other close contacts [10].
The clinical illness is characterized by a prodrome that can remain 5- 9 days before people seek medical assistance, which is a risk period for community contagion [11].
Also the asymptomatic transmission has been documented, and the viral load is not significantly different between symptomatic and asymptomatic people, therefore the infectiousness may be the same in asymptomatic cases [10, 12].
Once inside the airways, the S protein on the viral surface recognizes and sticks to the receptor protein called ACE2 and the virus attacks the ACE2-bearing cells lining the airways. It can infect upper as well as lower respiratory tracts and with the dying cells sloughing down and filling the airways, the virus is carried deeper into the lungs [13].
Significantly high ACE2 expression is found in type II alveolar cells (AT2) of the lung, upper esophageal stratified epithelial cells, absorptive enterocytes from ileum and colon, gall bladder and bile ductal cells, myocardial cells, kidney proximal tubule cells and bladder urothelial cells. Apart from this, the ACE2 is also expressed on the oral mucosa and highly enriched in epithelial cells of tongue accounting for a potentially high risk of this route for 2019-nCoV infectious susceptibility. As such, the ACE2 expression is higher in oral mucosa and tongue than buccal and gingival tissues. Further, the saliva, urine and stool specimens and rectal swabs have demonstrated embedded viruses in the COVID-19 patients1 [14, 15].
The WHO recommends collecting specimens from both the upper respiratory tract (nasopharyngeal and oropharyngeal samples) and lower respiratory tract such as expectorated sputum, endotracheal aspirate, or Broncho-alveolar lavage for a diagnostic test. The samples are needed to be stored at 4°C. The amplification of the genetic material extracted from the sample is done through a reverse polymerase chain reaction (RT-PCR) [16].
If the test result is positive, it is recommended that the test is to be repeated for verification. In patients with confirmed COVID-19 diagnosis, the laboratory evaluation should be repeated at intervals to evaluate for viral clearance prior to discharge from observation. In a positive case, lymphopenia as well as raised liver enzymes, LDH, muscle enzymes and C-reactive are negative prognostic factors [17].
Antibody tests are primarily used to determine if a person has already had covid-19. Specific IgM and IgG antibodies should start to become detectable after 4-5 days, with positive IgM antibodies in 70% of symptomatic patients by days 8-14 and 90% of total antibody tests positive by days 11-24. IgG reactivity is thought to reach >98% after several more weeks, but the duration of this antibody response is not yet known [16, 18-20].
Diagnosis of COVID-19 can also be performed using salivary diagnosis platforms [21]. Saliva samples could be collected in patients who present with oropharyngeal secretions as a symptom [22, 23]. Avoiding close contact between healthcare workers and infected patients to collect nasopharyngeal or oropharyngeal samples, the possibility of a saliva self-collection can strongly reduce the risk of COVID-19 transmission.
Besides, the nasopharyngeal and oropharyngeal collection promotes discomfort and may promote bleeding especially in infected patients with thrombocytopenia [24].
Furthermore, a disclosing virus test may be the rapidest and most practical way to test COVID-19.
In fact, we propose a new test technique based on the use of oral gel, mouthwash, or tablets that color the area where the virus is localized in mouth, and then COVID-19 infection can be diagnosed.
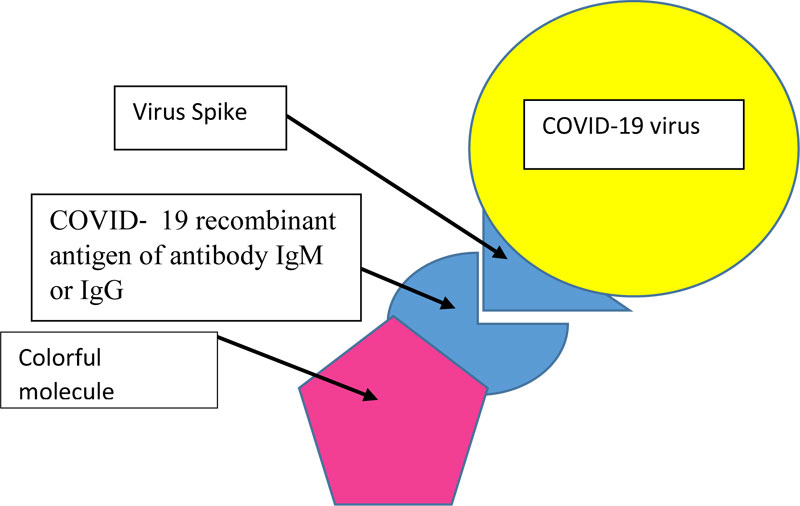
Actually, our test is composed of specific COVID-19 recombinant antigen of antibody IgM and IgG coupled to colorful or fluorescent molecules (Fig. 2). The person rinses his mouth with the mouthwash-test for 30 seconds, and rinses then with clean water. If we see some colored surfaces in the mouth, the subject is then COVID-19 positive. In case we use COVID-19 recombinant antigen of antibody IgG and IgM coupled to fluorescent molecules, we will see fluorescent surfaces in the mouth on ultraviolet light.
This disclosing test is very promising, because of its rapidity, low cost and easy use; prevents any contact with health workers, and also it will be more available than the other tests.
As of April 2020, the study is waiting to be funded and clinical trials will be prepared to be lunched to get advantage of the technique in order to improve COVID-19 testing.
AUTHOR’S CONTRIBUTIONS
Literature search, manuscript preparation and editing was done by R.A.A; manuscript review was carried out by A.B. The supervision was done by M.C and the whole manuscript was read and approved by all the authors.
CONFLICT OF INTEREST
The authors declare no conflicts of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The authors want to acknowledge the Editorial office of the journal and all the anonymous reviewers.


